Overview of these modules
The NIH-Wide Strategic Plan1 describes an integrative view of biology and human health that includes translational medicine, team science, and the importance of capitalizing on an exponentially growing and increasingly complex data ecosystem.2 Underlying this view is the need to use, share, and re-use biomedical data generated from widely varying experimental systems and researchers. Basic sources of biomedical data range from relatively small sets of measurements, such as animal body weights and bacterial cell counts that may be recorded by hand, to thousands or millions of instrument-generated data points from various imaging, -omic, and flow cytometry experiments. In either case, there is a generally common workflow that proceeds from measurement to data recording, pre-processing, analysis, and interpretation. However, in practice the distinct actions of data recording, data pre-processing, and data analysis are often merged or combined as a single entity by the researcher using commercial or open source spreadsheets, or as part of an often proprietary experimental measurement system / software combination (Figure 0.1), resulting in key failure points for reproducibility at the stages of data recording and pre-processing.
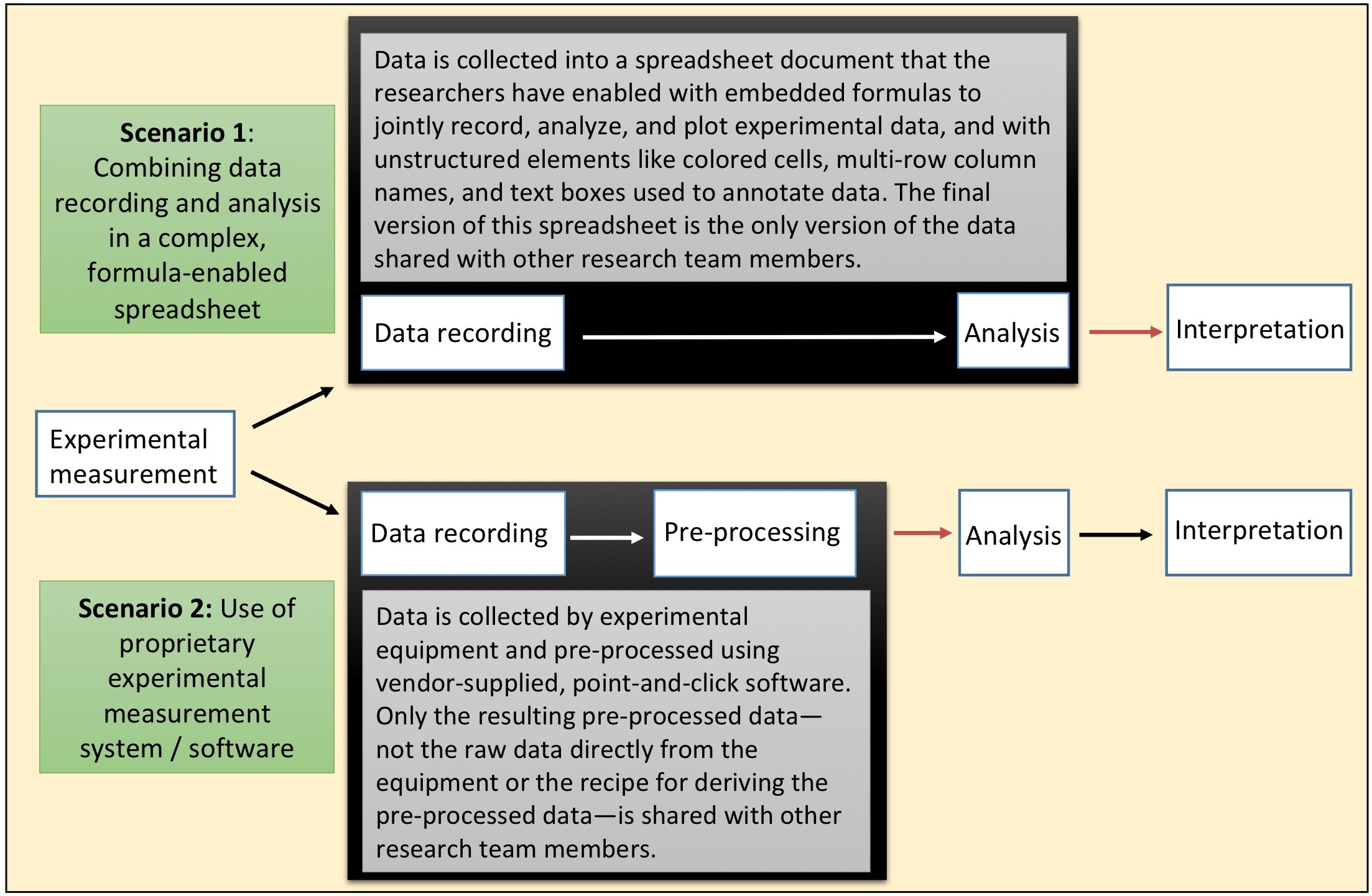
Figure 0.1: Two scenarios where ‘black boxes’ of non-transparent, non-reproducible data handling exist in research data workflows at the stages of data recording and pre-processing. These create potential points of failure for reproducible research. Red arrows indicate where data is passed to other research team members, including statisticians / data analysts, often within complex or unstructured spreadsheet files.
It is widely known and discussed among data scientists, mathematical modelers, and statisticians3 that there is frequently a need to discard, transform, and reformat various elements of the data shared with them by laboratory-based researchers, and that data is often shared in an unstructured format, increasing the risks of introducing errors through reformatting before applying more advanced computational methods. Instead, a critical need for reproducibility is for the transparent and clear sharing across research teams of: (1) raw data, directly from hand-recording or directly output from experimental equipment; (2) data that has been pre-processed as necessary (e.g., gating for flow cytometry data, feature identification for metabolomics data), saved in a consistent, structured format, and (3) a clear and repeatable description of how the pre-processed data was generated from the raw data.4
To enhance data reproducibility, it is critical to create a clear separation among data recording, data pre-processing, and data analysis—breaking up commonly existing ``black boxes” in data handling across the research process. Such a rigorous demarcation requires some change in the conventional understanding and use of spreadsheets and a recognition by biomedical researchers that recent advances in computer programming languages, especially the R programming language, provide user-friendly and accessible tools and concepts that can be used to extend a transparent and reproducible data workflow to the steps of data recording and pre-processing. Among our team, we have found that there are many common existing practices—including use of spreadsheets with embedded formulas that concurrently record and analyze experimental data, problematic management of project files, and reliance on proprietary, vendor-supplied point-and-click software for data pre-processing—that can interfere with the transparency, reproducibility, and efficiency of laboratory-based biomedical research projects, problems that have also been identified by others as key barriers to research reproducibility.5 In these training modules, we have choosen topics that tackle barriers to reproducibility that have straightforward, easy-to-teach solutions, but which are still very common in biomedical laboratory-based research programs.
License
This book is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, while all code in the book is under the MIT license.