Chapter 7 Exploring data #2
The video lectures for this chapter are embedded at relevant places in the text, with links to download a pdf of the associated slides for each video. You can also access a full playlist for the videos for this chapter.
7.1 Simple statistical tests in R
Download a pdf of the lecture slides for this video.
Download a pdf of the lecture slides for this video.
Let’s pull the fatal accident data just for the county that includes Las Vegas, NV. Each US county has a unique identifier (FIPS code), composed of a two-digit state FIPS and a three-digit county FIPS code. The state FIPS for Nevada is 32; the county FIPS for Clark County is 003. Therefore, we can filter down to Clark County data in the FARS data we collected with the following code:
library(readr)
library(dplyr)
clark_co_accidents <- read_csv("data/accident.csv") %>%
filter(STATE == 32 & COUNTY == 3)We can also check the number of accidents:
## # A tibble: 1 × 1
## n
## <int>
## 1 201We want to test if the probability, on a Friday or Saturday, of a fatal accident occurring is higher than on other days of the week. Let’s clean the data up a bit as a start:
library(tidyr)
library(lubridate)
clark_co_accidents <- clark_co_accidents %>%
select(DAY, MONTH, YEAR) %>%
unite(date, DAY, MONTH, YEAR, sep = "-") %>%
mutate(date = dmy(date))Here’s what the data looks like now:
## # A tibble: 5 × 1
## date
## <date>
## 1 2016-01-10
## 2 2016-02-21
## 3 2016-01-06
## 4 2016-01-13
## 5 2016-02-18Next, let’s get the count of accidents by date:
clark_co_accidents <- clark_co_accidents %>%
group_by(date) %>%
count() %>%
ungroup()
clark_co_accidents %>%
slice(1:3)## # A tibble: 3 × 2
## date n
## <date> <int>
## 1 2016-01-03 1
## 2 2016-01-06 1
## 3 2016-01-09 3We’re missing the dates without a fatal crash, so let’s add those. First, create a dataframe with all dates in 2016:
all_dates <- tibble(date = seq(ymd("2016-01-01"),
ymd("2016-12-31"), by = 1))
all_dates %>%
slice(1:5)## # A tibble: 5 × 1
## date
## <date>
## 1 2016-01-01
## 2 2016-01-02
## 3 2016-01-03
## 4 2016-01-04
## 5 2016-01-05Then merge this with the original dataset on Las Vegas fatal crashes and make any day missing from the fatal crashes dataset have a “0” for number of fatal accidents (n):
clark_co_accidents <- clark_co_accidents %>%
right_join(all_dates, by = "date") %>%
# If `n` is missing, set to 0. Otherwise keep value.
mutate(n = ifelse(is.na(n), 0, n))
clark_co_accidents %>%
slice(1:3)## # A tibble: 3 × 2
## date n
## <date> <dbl>
## 1 2016-01-03 1
## 2 2016-01-06 1
## 3 2016-01-09 3Next, let’s add some information about day of week and weekend:
clark_co_accidents <- clark_co_accidents %>%
mutate(weekday = wday(date, label = TRUE),
weekend = weekday %in% c("Fri", "Sat"))
clark_co_accidents %>%
slice(1:3)## # A tibble: 3 × 4
## date n weekday weekend
## <date> <dbl> <ord> <lgl>
## 1 2016-01-03 1 Sun FALSE
## 2 2016-01-06 1 Wed FALSE
## 3 2016-01-09 3 Sat TRUENow let’s calculate the probability that a day has at least one fatal crash, separately for weekends and weekdays:
clark_co_accidents <- clark_co_accidents %>%
mutate(any_crash = n > 0)
crash_prob <- clark_co_accidents %>%
group_by(weekend) %>%
summarize(n_days = n(),
crash_days = sum(any_crash)) %>%
mutate(prob_crash_day = crash_days / n_days)
crash_prob## # A tibble: 2 × 4
## weekend n_days crash_days prob_crash_day
## <lgl> <int> <int> <dbl>
## 1 FALSE 260 107 0.412
## 2 TRUE 106 43 0.406In R, you can use prop.test to test if two proportions are equal. Inputs include the total number of trials in each group (n =) and the number of “successes”” (x =):
##
## 2-sample test for equality of proportions with continuity correction
##
## data: crash_prob$crash_days out of crash_prob$n_days
## X-squared = 1.5978e-30, df = 1, p-value = 1
## alternative hypothesis: two.sided
## 95 percent confidence interval:
## -0.1109757 0.1227318
## sample estimates:
## prop 1 prop 2
## 0.4115385 0.4056604I won’t be teaching in this course how to find the correct statistical test. That’s something you’ll hopefully learn in a statistics course. There are also a variety of books that can help you with this, including some that you can access free online through CSU’s library. One servicable introduction is “Statistical Analysis with R for Dummies”.
You can create an object from the output of any statistical test in R. Typically, this will be (at least at some level) in an object class called a “list”:
## [1] TRUESo far, we’ve mostly worked with two object types in R, dataframes and vectors. In the next subsection we’ll look more at two object classes we haven’t looked at much, matrices and lists. Both have important roles once you start applying more advanced methods to analyze your data.
Download a pdf of the lecture slides for this video.
7.2 Matrices
A matrix is like a data frame, but all the values in all columns must be of the same class (e.g., numeric, character). R uses matrices a lot for its underlying math (e.g., for the linear algebra operations required for fitting regression models). R can do matrix operations quite quickly.
You can create a matrix with the matrix function. Input a vector with the values to fill the matrix and ncol to set the number of columns:
## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 3 5 7 9
## [2,] 2 4 6 8 10By default, the matrix will fill up by column. You can fill it by row with the byrow function:
## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 2 3 4 5
## [2,] 6 7 8 9 10In certain situations, you might want to work with a matrix instead of a data frame (for example, in cases where you were concerned about speed – a matrix is more memory efficient than the corresponding data frame). If you want to convert a data frame to a matrix, you can use the as.matrix function:
foo <- tibble(col_1 = 1:2, col_2 = 3:4,
col_3 = 5:6, col_4 = 7:8,
col_5 = 9:10)
(foo <- as.matrix(foo))## col_1 col_2 col_3 col_4 col_5
## [1,] 1 3 5 7 9
## [2,] 2 4 6 8 10You can index matrices with square brackets, just like data frames:
## col_1 col_2
## 1 3You cannot, however, use dplyr functions with matrices:
All elements in a matrix must have the same class.
The matrix will default to make all values the most general class of any of the values, in any column. For example, if we replaced one numeric value with the character “a”, everything would turn into a character:
## col_1 col_2 col_3 col_4 col_5
## [1,] "a" "3" "5" "7" "9"
## [2,] "2" "4" "6" "8" "10"7.3 Lists
A list has different elements, just like a data frame has different columns. However, the different elements of a list can have different lengths (unlike the columns of a data frame). The different elements can also have different classes.
bar <- list(some_letters = letters[1:3],
some_numbers = 1:5,
some_logical_values = c(TRUE, FALSE))
bar## $some_letters
## [1] "a" "b" "c"
##
## $some_numbers
## [1] 1 2 3 4 5
##
## $some_logical_values
## [1] TRUE FALSETo index an element from a list, use double square brackets. You can use bracket indexing either with numbers (which element in the list?) or with names:
## [1] "a" "b" "c"## [1] 1 2 3 4 5You can also index lists with the $ operator:
## [1] TRUE FALSETo access a specific value within a list element we can index the element e.g.:
## [1] "b"Lists can be used to contain data with an unusual structure and / or lots of different components. For example, the information from fitting a regression is often stored as a list:
## [1] TRUEThe names function returns the name of each element in the list:
## [1] "coefficients" "residuals" "fitted.values"## (Intercept) c(1:10)
## 0.44067097 -0.07168315A list can even contain other lists. We can use the str function to see the structure of a list:
## List of 2
## $ :List of 2
## ..$ : chr "a"
## ..$ : chr "b"
## $ :List of 2
## ..$ : num 1
## ..$ : num 2Sometimes you’ll see unnecessary lists-of-lists, perhaps when importing data into R created. Or a list with multiple elements that you would like to combine. You can remove a level of hierarchy from a list using the flatten function from the purrr package:
## [[1]]
## [[1]][[1]]
## [1] "a"
##
## [[1]][[2]]
## [1] "b"
##
##
## [[2]]
## [[2]][[1]]
## [1] 1
##
## [[2]][[2]]
## [1] 2## [[1]]
## [1] "a"
##
## [[2]]
## [1] "b"
##
## [[3]]
## [1] 1
##
## [[4]]
## [1] 2Let’s look at the list object from the statistical test we ran for Las Vegas:
## List of 9
## $ statistic : Named num 1.6e-30
## ..- attr(*, "names")= chr "X-squared"
## $ parameter : Named num 1
## ..- attr(*, "names")= chr "df"
## $ p.value : num 1
## $ estimate : Named num [1:2] 0.412 0.406
## ..- attr(*, "names")= chr [1:2] "prop 1" "prop 2"
## $ null.value : NULL
## $ conf.int : num [1:2] -0.111 0.123
## ..- attr(*, "conf.level")= num 0.95
## $ alternative: chr "two.sided"
## $ method : chr "2-sample test for equality of proportions with continuity correction"
## $ data.name : chr "crash_prob$crash_days out of crash_prob$n_days"
## - attr(*, "class")= chr "htest"Using str to print out the list’s structure doesn’t produce the easiest to digest output. We can use the jsonedit function from the listviewer package to create a widget in the viewer pane to more esily explore our list.
We can pull out an element using the $ notation:
## [1] 1Or using the [[ notation:
## prop 1 prop 2
## 0.4115385 0.4056604You may have noticed, though, that this output is not a tidy dataframe.
Ack! That means we can’t use all the tidyverse tricks we’ve learned so far in the course!
Fortunately, David Robinson noticed this problem and came up with a package called broom that can “tidy up” a lot of these kinds of objects.
The broom package has three main functions:
glance: Return a one-row, tidy dataframe from a model or other R objecttidy: Return a tidy dataframe from a model or other R objectaugment: “Augment” the dataframe you input to the statistical function
Here is the output for tidy for the vegas_test object (augment
won’t work for this type of object, and glance gives the same thing as tidy):
## # A tibble: 1 × 9
## estimate1 estimate2 statistic p.value parameter conf.low conf.high method
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 0.412 0.406 1.60e-30 1.000 1 -0.111 0.123 2-sample t…
## # ℹ 1 more variable: alternative <chr>7.4 Apply a test multiple times
Download a pdf of the lecture slides for this video.
Download a pdf of the lecture slides for this video.
Download a pdf of the lecture slides for this video.
Often, we don’t want to just apply a statistical test to our entire data set.
Let’s look at an example from the microbiome package.
## [1] "lipids" "microbes" "meta" "phyloseq"Like we saw before, the list-like phyloseq objects require a little tidying before we can use them easiliy.
library(dplyr)
peerj32_tibble <- (psmelt(peerj32$phyloseq)) %>%
dplyr::select(-Sample) %>%
rename_all(tolower) %>%
as_tibble()## # A tibble: 3 × 10
## otu abundance time sex subject sample group phylum family genus
## <chr> <dbl> <int> <fct> <fct> <chr> <fct> <chr> <chr> <chr>
## 1 Bacteroides vu… 9734 1 male S19 sampl… LGG Bacte… Bacte… Bact…
## 2 Bacteroides vu… 5883 1 fema… S14 sampl… Plac… Bacte… Bacte… Bact…
## 3 Bacteroides vu… 5134 2 fema… S8 sampl… LGG Bacte… Bacte… Bact…We can make a plot from the resulting tibble to help us explore the data.
library(ggplot2)
ggplot(peerj32_tibble, aes(x = subject, y = abundance, color = sex)) +
geom_point() +
theme_bw() +
facet_wrap(~phylum, ncol = 4, scales = "free") +
theme(axis.text.x = element_blank(),
legend.position = "top") +
xlab("Subject ID")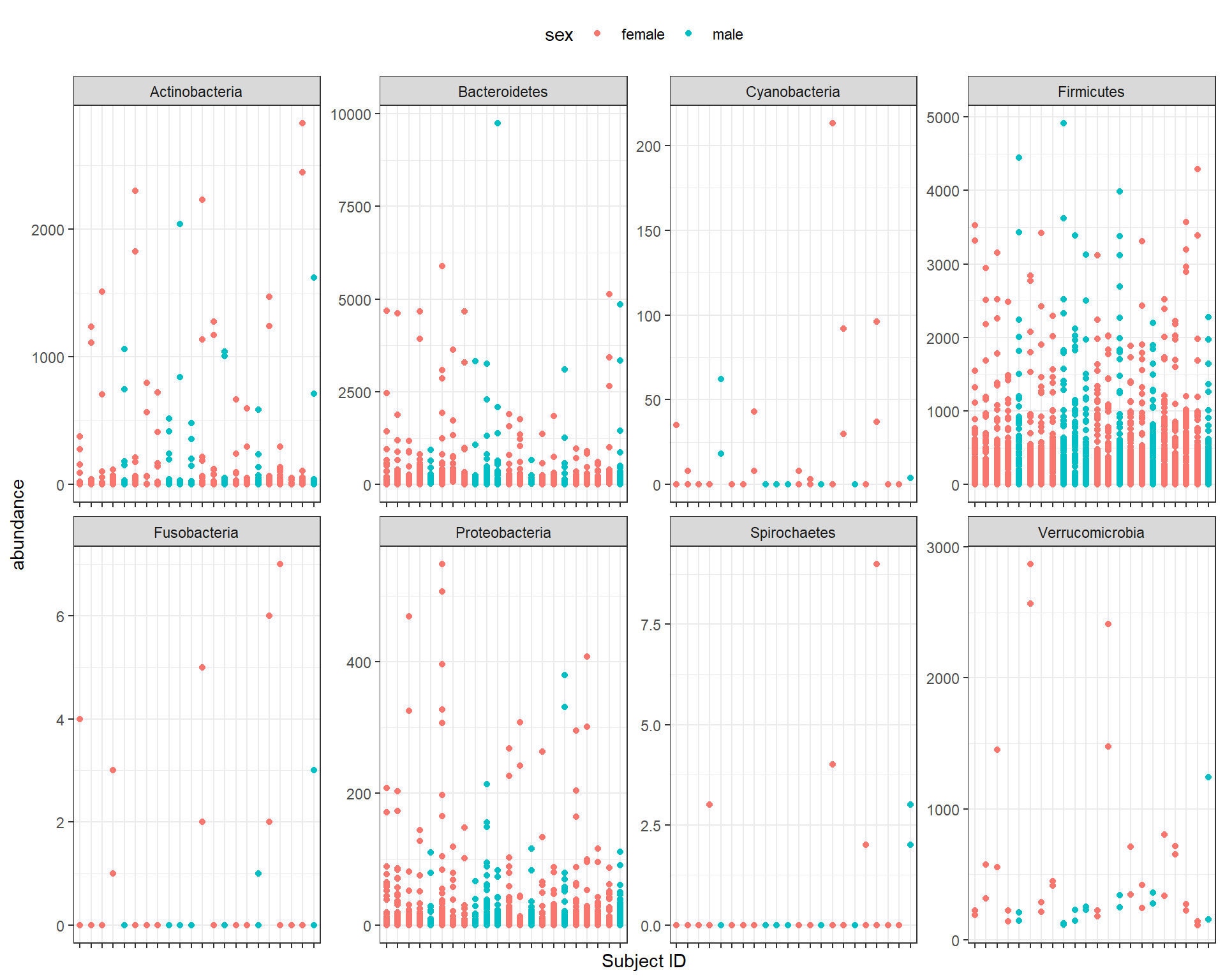
We can group the data by phylum and group and then use the nest function from the tidyr package to create a new dataframe containing the nested data:
## # A tibble: 16 × 3
## # Groups: phylum, group [16]
## group phylum data
## <fct> <chr> <list>
## 1 LGG Actinobacteria <tibble [128 × 8]>
## 2 Placebo Actinobacteria <tibble [224 × 8]>
## 3 LGG Bacteroidetes <tibble [256 × 8]>
## 4 Placebo Bacteroidetes <tibble [448 × 8]>
## 5 LGG Cyanobacteria <tibble [16 × 8]>
## 6 Placebo Cyanobacteria <tibble [28 × 8]>
## 7 LGG Firmicutes <tibble [1,216 × 8]>
## 8 Placebo Firmicutes <tibble [2,128 × 8]>
## 9 LGG Fusobacteria <tibble [16 × 8]>
## 10 Placebo Fusobacteria <tibble [28 × 8]>
## 11 LGG Proteobacteria <tibble [416 × 8]>
## 12 Placebo Proteobacteria <tibble [728 × 8]>
## 13 LGG Spirochaetes <tibble [16 × 8]>
## 14 Placebo Spirochaetes <tibble [28 × 8]>
## 15 LGG Verrucomicrobia <tibble [16 × 8]>
## 16 Placebo Verrucomicrobia <tibble [28 × 8]>We can then use the map function from the purrr package to apply functions to each nested dataframe. Let’s start by counting the rows in each nested dataframe and filtering out dataframes with less than 25 rows.
library(purrr)
filtered_data <- nested_data %>%
mutate(n_rows = purrr::map(data, ~ nrow(.x))) %>%
filter(n_rows > 25)Now let’s perform a t-test on each out the nested dataframes. Remember, each nested dataframe is one unique combination of phylum and group.
## # A tibble: 12 × 5
## # Groups: phylum, group [12]
## group phylum data n_rows t_test
## <fct> <chr> <list> <list> <list>
## 1 LGG Bacteroidetes <tibble [256 × 8]> <int [1]> <htest>
## 2 Placebo Bacteroidetes <tibble [448 × 8]> <int [1]> <htest>
## 3 Placebo Firmicutes <tibble [2,128 × 8]> <int [1]> <htest>
## 4 LGG Firmicutes <tibble [1,216 × 8]> <int [1]> <htest>
## 5 Placebo Verrucomicrobia <tibble [28 × 8]> <int [1]> <htest>
## 6 LGG Actinobacteria <tibble [128 × 8]> <int [1]> <htest>
## 7 Placebo Actinobacteria <tibble [224 × 8]> <int [1]> <htest>
## 8 Placebo Proteobacteria <tibble [728 × 8]> <int [1]> <htest>
## 9 LGG Proteobacteria <tibble [416 × 8]> <int [1]> <htest>
## 10 Placebo Cyanobacteria <tibble [28 × 8]> <int [1]> <htest>
## 11 Placebo Fusobacteria <tibble [28 × 8]> <int [1]> <htest>
## 12 Placebo Spirochaetes <tibble [28 × 8]> <int [1]> <htest>The resulting dataframe contains a new column, which contains the model objects. Just as we mapped the t.test function, we can map the tidying functions from the broom package to extract the information we want from the nested model objects.
## # A tibble: 12 × 6
## # Groups: phylum, group [12]
## group phylum data n_rows t_test summary
## <fct> <chr> <list> <list> <list> <list>
## 1 LGG Bacteroidetes <tibble [256 × 8]> <int [1]> <htest> <tibble>
## 2 Placebo Bacteroidetes <tibble [448 × 8]> <int [1]> <htest> <tibble>
## 3 Placebo Firmicutes <tibble [2,128 × 8]> <int [1]> <htest> <tibble>
## 4 LGG Firmicutes <tibble [1,216 × 8]> <int [1]> <htest> <tibble>
## 5 Placebo Verrucomicrobia <tibble [28 × 8]> <int [1]> <htest> <tibble>
## 6 LGG Actinobacteria <tibble [128 × 8]> <int [1]> <htest> <tibble>
## 7 Placebo Actinobacteria <tibble [224 × 8]> <int [1]> <htest> <tibble>
## 8 Placebo Proteobacteria <tibble [728 × 8]> <int [1]> <htest> <tibble>
## 9 LGG Proteobacteria <tibble [416 × 8]> <int [1]> <htest> <tibble>
## 10 Placebo Cyanobacteria <tibble [28 × 8]> <int [1]> <htest> <tibble>
## 11 Placebo Fusobacteria <tibble [28 × 8]> <int [1]> <htest> <tibble>
## 12 Placebo Spirochaetes <tibble [28 × 8]> <int [1]> <htest> <tibble>The final step is to return a tidy dataframe, we can do this using the unnest function from the tidyr package. Note: it is also wise to ungroup after you are done operating on your grouped variables - you may run into problems if you forget your dataframe is grouped later on!
## # A tibble: 12 × 3
## group phylum p.value
## <fct> <chr> <dbl>
## 1 LGG Bacteroidetes 0.930
## 2 Placebo Bacteroidetes 0.828
## 3 Placebo Firmicutes 0.344
## 4 LGG Firmicutes 0.131
## 5 Placebo Verrucomicrobia 0.106
## 6 LGG Actinobacteria 0.600
## 7 Placebo Actinobacteria 0.997
## 8 Placebo Proteobacteria 0.856
## 9 LGG Proteobacteria 0.0111
## 10 Placebo Cyanobacteria 0.193
## 11 Placebo Fusobacteria 0.265
## 12 Placebo Spirochaetes 0.728We can then plot the results. Let’s do this for the “LGG” (non-placebo) group, using our principles for good graphics.
p_data <- unnested %>%
filter(group == "LGG") %>%
mutate(phylum = fct_reorder(phylum, p.value))
ggplot(p_data, aes(y = p.value, x = phylum)) +
facet_wrap(~group) +
geom_bar(stat="identity", fill = "grey90", color = "grey20") +
coord_flip() +
theme_bw() +
ggtitle("Difference in abundance by gender") +
xlab("") + ylab("p-value from t-test")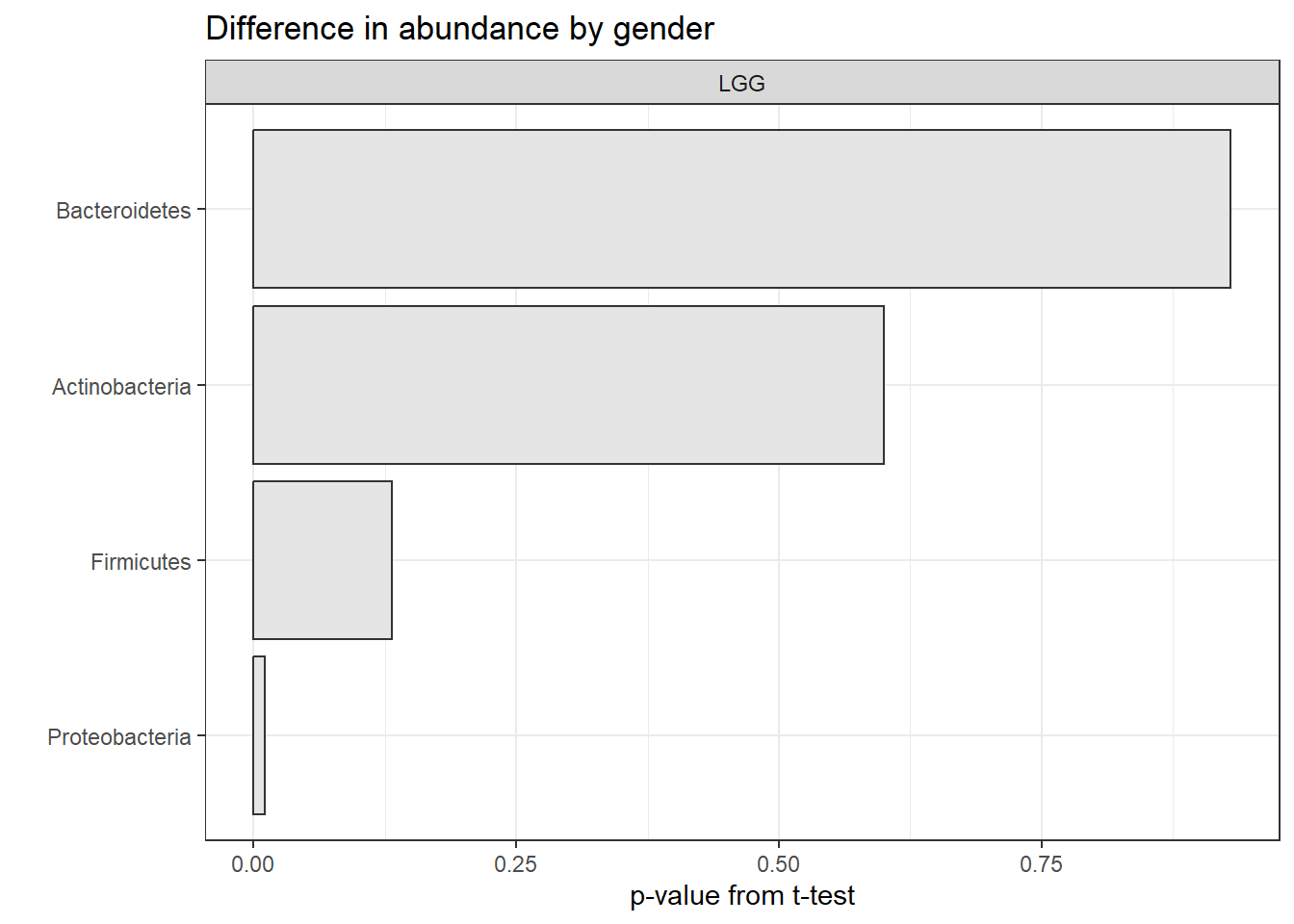
7.5 Regression models
Download a pdf of the lecture slides for this video.
Download a pdf of the lecture slides for this video.
7.5.1 Formula structure
Regression models can be used to estimate how the expected value of a dependent variable changes as independent variables change.
In R, regression formulas take this structure:
Notice that a tilde, ~, is used to separate the independent and dependent variables and that a plus sign, +, is used to join independent variables. This format mimics the statistical notation:
\[ Y_i \sim X_1 + X_2 + X_3 \]
You will use this type of structure in R fo a lot of different function calls, including those for linear models (fit with the lm function) and generalized linear models (fit with the glm function).
There are some conventions that can be used in R formulas. Common ones include:
| Convention | Meaning |
|---|---|
I() |
evaluate the formula inside I() before fitting (e.g.,
I(x1 + x2)) |
: |
fit the interaction between x1 and x2 variables |
* |
fit the main effects and interaction for both variables
(e.g., x1*x2 equals x1 + x2 + x1:x2) |
. |
include as independent variables all variables other than
the response (e.g., y ~ .) |
1 |
intercept (e.g., y ~ 1 for an intercept-only model) |
- |
do not include a variable in the data frame as an
independent variables (e.g., y ~ . - x1); usually used
in conjunction with . or 1 |
7.5.2 Linear models
To fit a linear model, you can use the function lm(). This function is part of the stats package, which comes installed with base R. In this function, you can use the data option to specify the data frame from which to get the vectors.
## Warning: There was 1 warning in `mutate()`.
## ℹ In argument: `sex = fct_recode(factor(sex), Male = "1", Female = "2")`.
## Caused by warning:
## ! Unknown levels in `f`: 1, 2This previous call fits the model:
\[ Y_{i} = \beta_{0} + \beta_{1}X_{1,i} + \epsilon_{i} \]
where:
- \(Y_{i}\) : weight of child \(i\)
- \(X_{1,i}\) : height of child \(i\)
If you run the lm function without saving it as an object, R will fit the regression and print out the function call and the estimated model coefficients:
##
## Call:
## lm(formula = wt ~ ht, data = nepali)
##
## Coefficients:
## (Intercept) ht
## -8.7581 0.2342However, to be able to use the model later for things like predictions and model assessments, you should save the output of the function as an R object:
This object has a special class, lm:
## [1] "lm"This class is a special type of list object. If you use is.list to check, you can confirm that this object is a list:
## [1] TRUEThere are a number of functions that you can apply to an lm object. These include:
| Function | Description |
|---|---|
summary |
Get a variety of information on the model, including coefficients and p-values for the coefficients |
coefficients |
Pull out just the coefficients for a model |
fitted |
Get the fitted values from the model (for the data used to fit the model) |
plot |
Create plots to help assess model assumptions |
residuals |
Get the model residuals |
For example, you can get the coefficients from the model by running:
## (Intercept) ht
## -8.7581466 0.2341969The estimated coefficient for the intercept is always given under the name “(Intercept)”. Estimated coefficients for independent variables are given based on their column names in the original data (“ht” here, for \(\beta_1\), or the estimated increase in expected weight for a one unit increase in height).
You can use the output from a coefficients call to plot a regression line based on the model fit on top of points showing the original data (Figure 7.1).
mod_coef <- coefficients(mod_a)
ggplot(nepali, aes(x = ht, y = wt)) +
geom_point(size = 0.2) +
xlab("Height (cm)") + ylab("Weight (kg)") +
geom_abline(aes(intercept = mod_coef[1],
slope = mod_coef[2]), col = "blue")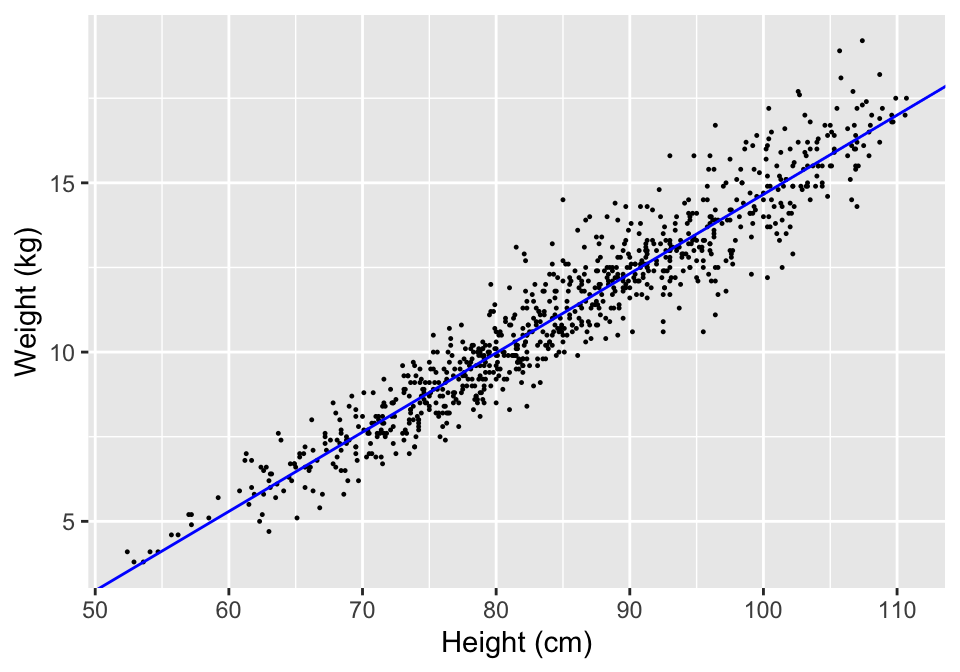
Figure 7.1: Example of using the output from a coefficients call to add a regression line to a scatterplot.
You can also add a linear regression line to a scatterplot by adding
the geom geom_smooth using the argument
method = “lm”.
You can use the function residuals on an lm object to pull out the residuals from the model fit:
## 1 2 3 4 6 7
## 0.1993922 -0.4329393 -0.4373953 -0.1355300 -0.6749080 -1.0838199The result of a residuals call is a vector with one element for each of the non-missing observations (rows) in the data frame you used to fit the model. Each value gives the different between the model fitted value and the observed value for each of these observations, in the same order the observations show up in the data frame. The residuals are in the same order as the observations in the original data frame.
You can also use the shorter function coef as an
alternative to coefficients and the shorter function
resid as an alternative to residuals.
As noted in the subsection on simple statistics functions, the summary function returns different output depending on the type of object that is input to the function. If you input a regression model object to summary, the function gives you a lot of information about the model. For example, here is the output returned by running summary for the linear regression model object we just created:
##
## Call:
## lm(formula = wt ~ ht, data = nepali)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.0077 -0.5479 -0.0293 0.4972 3.3514
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.758147 0.211529 -41.40 <2e-16 ***
## ht 0.234197 0.002459 95.23 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.8733 on 875 degrees of freedom
## (123 observations deleted due to missingness)
## Multiple R-squared: 0.912, Adjusted R-squared: 0.9119
## F-statistic: 9068 on 1 and 875 DF, p-value: < 2.2e-16This output includes a lot of useful elements, including (1) basic summary statistics for the residuals (to meet model assumptions, the median should be around zero and the absolute values fairly similar for the first and third quantiles), (2) coefficient estimates, standard errors, and p-values, and (3) some model summary statistics, including residual standard error, degrees of freedom, number of missing observations, and F-statistic.
The object returned by the summary() function when it is applied to an lm object is a list, which you can confirm using the is.list function:
## [1] TRUEWith any list, you can use the names function to get the names of all of the different elements of the object:
## [1] "call" "terms" "residuals" "coefficients"
## [5] "aliased" "sigma" "df" "r.squared"
## [9] "adj.r.squared" "fstatistic" "cov.unscaled" "na.action"You can use the $ operator to pull out any element of the list. For example, to pull out the table with information on the estimated model coefficients, you can run:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.7581466 0.211529182 -41.40396 2.411051e-208
## ht 0.2341969 0.002459347 95.22726 0.000000e+00The plot function, like the summary function, will give different output depending on the class of the object that you input. For an lm object, you can use the plot function to get a number of useful diagnostic plots that will help you check regression assumptions (Figure 7.2):
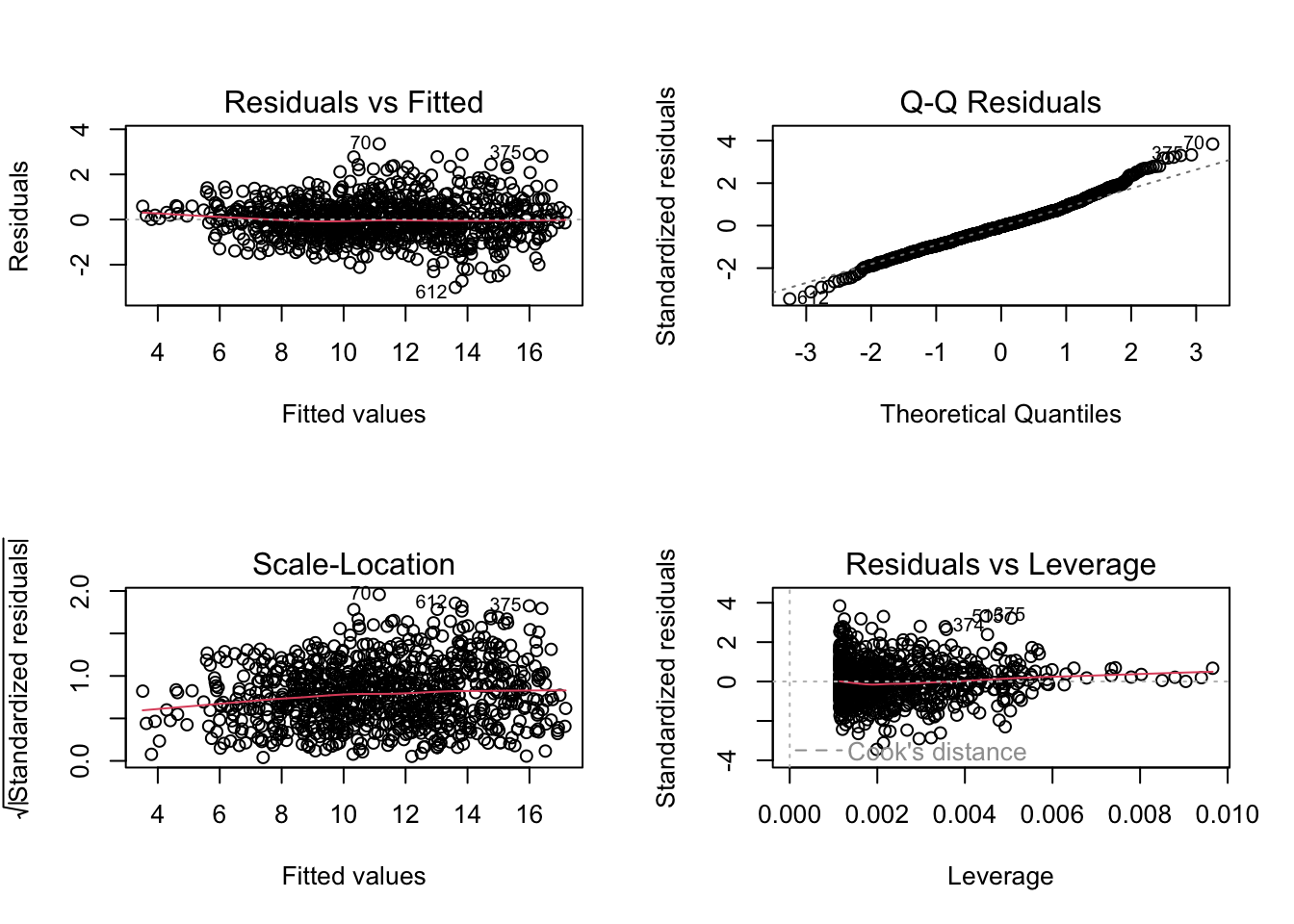
Figure 7.2: Example output from running the plot function with an lm object as the input.
You can also use binary variables or factors as independent variables in regression models. For example, in the nepali dataset, sex is a factor variable with the levels “Male” and “Female”. You can fit a linear model of weight regressed on sex for this data with the call:
This call fits the model:
\[ Y_{i} = \beta_{0} + \beta_{1}X_{1,i} + \epsilon_{i} \]
where \(X_{1,i}\) : sex of child \(i\), where 0 = male and 1 = female.
Here are the estimated coefficients from fitting this model:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 11.4226374 0.1375427 83.047920 0.00000000
## sexFemale -0.4866184 0.1982810 -2.454185 0.01431429You’ll notice that, in addition to an estimated intercept ((Intercept)), the other estimated coefficient is sexFemale rather than just sex, although the column name in the data frame input to lm for this variable is sex.
This is because, when a factor or binary variable is input as an independent variable in a linear regression model, R will fit an estimated coefficient for all levels of factors except the first factor level. By default, this first factor level is used as the baseline level, and so its estimated mean is given by the estimated intercept, while the other model coefficients give the estimated difference from this baseline.
For example, the model fit above tells us that the estimated mean weight of males is 11.4, while the estimated mean weight of females is 11.4 + -0.5 = 10.9.
7.6 Handling model objects
The broom package contains tools for converting statistical objects into nice tidy data frames. The tools in the broom package make it easy to process statistical results in R using the tools of the tidyverse.
7.6.1 broom::tidy
The tidy() function returns a data frame with information on the fitted model terms. For example, when applied to one of our linear models we get:
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -8.758 | 0.212 | -41.404 | 0 |
| ht | 0.234 | 0.002 | 95.227 | 0 |
## [1] "tbl_df" "tbl" "data.frame"You can pass arguments to the tidy() function. For example, include confidence intervals:
| term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|---|---|---|---|---|---|
| (Intercept) | -8.758 | 0.212 | -41.404 | 0 | -9.173 | -8.343 |
| ht | 0.234 | 0.002 | 95.227 | 0 | 0.229 | 0.239 |
7.6.2 broom::augment
The augment() function adds information about a fitted model to the dataset used to fit the model. For example, when applied to one of our linear models we get information on the fitted values and residuals included in the output:
| .rownames | wt | ht | .fitted | .resid | .hat | .sigma | .cooksd | .std.resid |
|---|---|---|---|---|---|---|---|---|
| 1 | 12.8 | 91.2 | 12.601 | 0.199 | 0.001 | 0.874 | 0 | 0.228 |
| 2 | 12.8 | 93.9 | 13.233 | -0.433 | 0.002 | 0.874 | 0 | -0.496 |
| 3 | 13.1 | 95.2 | 13.537 | -0.437 | 0.002 | 0.874 | 0 | -0.501 |
7.6.3 broom::glance
The glance() functions returns a one row summary of a fitted model object: For example:
| r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC | deviance | df.residual | nobs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.912 | 0.912 | 0.873 | 9068.23 | 0 | 1 | -1124.615 | 2255.23 | 2269.56 | 667.351 | 875 | 877 |
7.6.4 References– statistics in R
One great (and free online for CSU students through our library) book to find out more about using R for basic statistics is:
If you want all the details about fitting linear models and GLMs in R, Julian Faraway’s books are fantastic. He has one on linear models and one on extensions including logistic and Poisson models:
- Linear Models with R (also free online through the CSU library)
- Extending the Linear Model with R
7.7 Functions
Download a pdf of the lecture slides for this video.
Download a pdf of the lecture slides for this video.
Download a pdf of the lecture slides for this video.
As you move to larger projects, you will find yourself using the same code a lot.
Examples include:
- Reading in data from a specific type of equipment (air pollution monitor, accelerometer)
- Running a specific type of analysis
- Creating a specific type of plot or map
If you find yourself cutting and pasting a lot, convert the code to a function.
Advantages of writing functions include:
- Coding is more efficient
- Easier to change your code (if you’ve cut and paste code and you want to change something, you have to change it everywhere - this is an easy way to accidentally create bugs in your code)
- Easier to share code with others
You can name a function anything you want (although try to avoid names of preexisting-existing functions). You then define any inputs (arguments; separate multiple arguments with commas) and put the code to run in braces:
Here is an example of a very basic function. This function takes a number as input and adds 1 to that number.
## [1] 4## [1] 0- Functions can input any type of R object (for example, vectors, data frames, even other functions and ggplot objects)
- Similarly, functions can output any type of R object
- When defining a function, you can set default values for some of the parameters
- You can explicitly specify the value to return from the function
For example, the following function inputs a data frame (datafr) and a one-element vector (child_id) and returns only rows in the data frame where it’s id column matches child_id. It includes a default value for datafr, but not for child_id.
subset_nepali <- function(datafr = nepali, child_id){
datafr <- datafr %>%
filter(id == child_id)
return(datafr)
}If an argument is not given for a parameter with a default, the function will run using the default value for that parameter. For example:
## id sex wt ht mage lit died alive age
## 1 120011 Male 12.8 91.2 35 0 2 5 41
## 2 120011 Male 12.8 93.9 35 0 2 5 45
## 3 120011 Male 13.1 95.2 35 0 2 5 49
## 4 120011 Male 13.8 96.9 35 0 2 5 53
## 5 120011 Male NA NA 35 0 2 5 57If an argument is not given for a parameter without a default, the function call will result in an error. For example:
## Error in `filter()`:
## ℹ In argument: `id == child_id`.
## Caused by error:
## ! argument "child_id" is missing, with no defaultBy default, the function will return the last defined object, although the choice of using return can affect printing behavior when you run the function. For example, I could have written the subset_nepali function like this:
In this case, the output will not automatically print out when you call the function without assigning it to an R object:
However, the output can be assigned to an R object in the same way as when the function was defined without return:
## id sex wt ht mage lit died alive age
## 1 120011 Male 12.8 91.2 35 0 2 5 41
## 2 120011 Male 12.8 93.9 35 0 2 5 45
## 3 120011 Male 13.1 95.2 35 0 2 5 49
## 4 120011 Male 13.8 96.9 35 0 2 5 53
## 5 120011 Male NA NA 35 0 2 5 57R is very “good” at running functions! It will look for (scope) the variables in your function in various places (environments). So your functions may run even when you don’t expect them to, potentially, with unexpected results!
The return function can also be used to return an object other than the last defined object (although this doesn’t tend to be something you need to do very often). If you did not use return in the following code, it will output “Test output”:
subset_nepali <- function(datafr = nepali, child_id){
datafr <- datafr %>%
filter(id == child_id)
a <- "Test output"
}
(subset_nepali(child_id = "120011"))## [1] "Test output"Conversely, you can use return to output datafr, even though it’s not the last object defined:
subset_nepali <- function(datafr = nepali, child_id){
datafr <- datafr %>%
filter(id == child_id)
a <- "Test output"
return(datafr)
}
subset_nepali(child_id = "120011")## id sex wt ht mage lit died alive age
## 1 120011 Male 12.8 91.2 35 0 2 5 41
## 2 120011 Male 12.8 93.9 35 0 2 5 45
## 3 120011 Male 13.1 95.2 35 0 2 5 49
## 4 120011 Male 13.8 96.9 35 0 2 5 53
## 5 120011 Male NA NA 35 0 2 5 577.7.1 if / else statements
There are other control structures you can use in your R code. Two that you will commonly use within R functions are if and ifelse statements.
An if statement tells R that, if a certain condition is true, do run some code. For example, if you wanted to print out only odd numbers between 1 and 5, one way to do that is with an if statement: (Note: the %% operator in R returns the remainder of the first value (i) divided by the second value (2).)
## [1] 1
## [1] 3
## [1] 5The if statement runs some code if a condition is true, but does nothing if it is false. If you’d like different code to run depending on whether the condition is true or false, you can us an if / else or an if / else if / else statement.
## [1] 1
## [1] "2 is even"
## [1] 3
## [1] "4 is even"
## [1] 5What would this code do?
for(i in 1:100){
if(i %% 3 == 0 & i %% 5 == 0){
print("FizzBuzz")
} else if(i %% 3 == 0){
print("Fizz")
} else if(i %% 5 == 0){
print("Buzz")
} else {
print(i)
}
}If / else statements are extremely useful in functions.
In R, the if statement evaluates everything in the parentheses and, if that evaluates to TRUE, runs everything in the braces. This means that you can trigger code in an if statement with a single-value logical vector:
## [1] "It's the weekend!"This functionality can be useful with parameters you choose to include when writing your own functions (e.g., print = TRUE).
7.7.2 Some other control structures
The control structure you are most likely to use in data analysis with R is the “if / else” statement. However, there are a few other control structures you may occasionally find useful:
nextbreakwhile
You can use the next structure to skip to the next round of a loop when a certain condition is met. For example, we could have used this code to print out odd numbers between 1 and 5:
## [1] 1
## [1] 3
## [1] 5You can use break to break out of a loop if a certain condition is met. For example, the final code will break out of the loop once i is over 2, so it will only print the numbers 1 through 3:
## [1] 1
## [1] 2
## [1] 37.8 In-course exercise for Chapter 7
7.8.1 Exploring taxonomic profiling data
- We’ll be using a package on Bioconductor called
microbiome. You’ll need to install that package from Bioconductor. This uses code that’s different from the default you use to download a package from CRAN. Go to the Bioconductor page for the microbiome package and figure out how to install this package based on instructions on that page. - The
microbiomethat includes tools for exploring and analysing microbiome profiling data. This package has a website with tutorial information here. We want to explore a dataset on genus-level microbiota profiling (atlas1006). Navigate to the tutorial webpage to figure out how you can get this example raw data loaded in your R session. Use theclassandstrfunctions to start exploring this data. Is it in a dataframe (tibble)? Is it in a tidy format? How is the data structured? - On the microbiome page, find the documentation describing the
atlas1006data. Look through this documentation to figure out what information is included in the data. Also, check the helpfile for this dataset and look up the original article describing the data (you can find the article information in the help resources). - The
atlas1006data is stored in a special object class called a “phyloseq” object (you should have seen this when you usedclasswith the object). You can pull certain parts of this data using special functions called “accessors”. One isget_variable. Try runningget_variablewith theatlas1006data. What do you think this data is showing? - Which different nationalities are represented by the study subjects, based on the dataframe you extracted in the last step? How many samples have each nationality? Which different BMI groups are included? Does it look like the study was balanced among these groups?
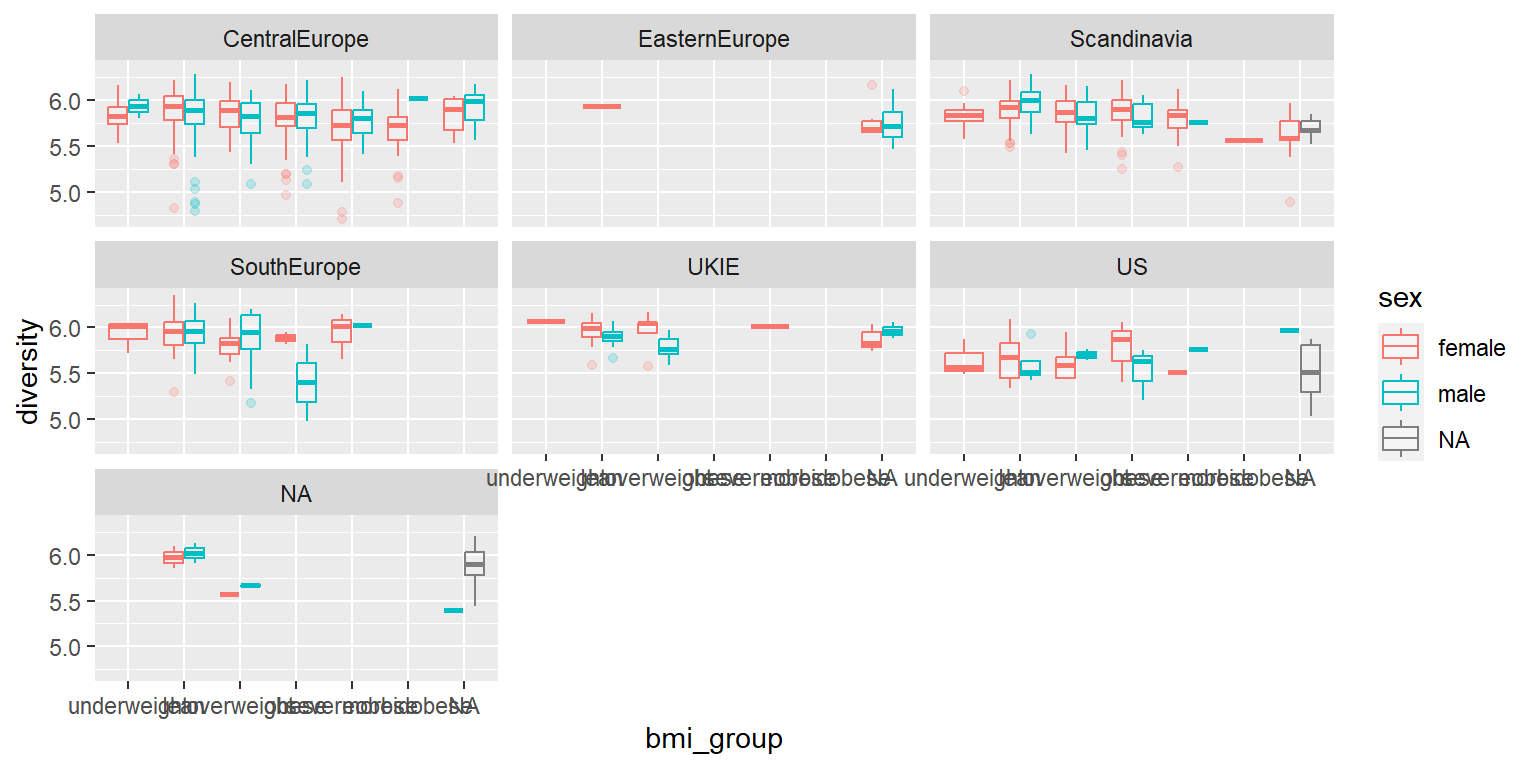
- Based on the data you extracted, does it look like diversity varies much between males and females? Across BMI groups?
- Discuss what steps you would need to take to create the following plot. To start, don’t write any code, just develop a plan. Talk about what the dataset should look like right before you create the plot and what functions you could use to get the data from its current format to that format.
- Try to write the code to create this plot. This will include some code for cleaning the data and some code for plotting. I will add one example answer after class, but I’d like you to try to figure it out yourselves first.
7.8.1.1 Example R code
Install the microbiome package from Bioconductor:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("microbiome")Load the atlas1006 example data in the microbiome package and explore it with str and class:
## [1] "phyloseq"
## attr(,"package")
## [1] "phyloseq"## Formal class 'phyloseq' [package "phyloseq"] with 5 slots
## ..@ otu_table:Formal class 'otu_table' [package "phyloseq"] with 2 slots
## .. .. ..@ .Data : num [1:130, 1:1151] 0 0 0 21 1 72 0 0 176 10 ...
## .. .. .. ..- attr(*, "dimnames")=List of 2
## .. .. .. .. ..$ : chr [1:130] "Actinomycetaceae" "Aerococcus" "Aeromonas" "Akkermansia" ...
## .. .. .. .. ..$ : chr [1:1151] "Sample-1" "Sample-2" "Sample-3" "Sample-4" ...
## .. .. ..@ taxa_are_rows: logi TRUE
## .. .. ..$ dim : int [1:2] 130 1151
## .. .. ..$ dimnames:List of 2
## .. .. .. ..$ : chr [1:130] "Actinomycetaceae" "Aerococcus" "Aeromonas" "Akkermansia" ...
## .. .. .. ..$ : chr [1:1151] "Sample-1" "Sample-2" "Sample-3" "Sample-4" ...
## ..@ tax_table:Formal class 'taxonomyTable' [package "phyloseq"] with 1 slot
## .. .. ..@ .Data: chr [1:130, 1:3] "Actinobacteria" "Firmicutes" "Proteobacteria" "Verrucomicrobia" ...
## .. .. .. ..- attr(*, "dimnames")=List of 2
## .. .. .. .. ..$ : chr [1:130] "Actinomycetaceae" "Aerococcus" "Aeromonas" "Akkermansia" ...
## .. .. .. .. ..$ : chr [1:3] "Phylum" "Family" "Genus"
## .. .. ..$ dim : int [1:2] 130 3
## .. .. ..$ dimnames:List of 2
## .. .. .. ..$ : chr [1:130] "Actinomycetaceae" "Aerococcus" "Aeromonas" "Akkermansia" ...
## .. .. .. ..$ : chr [1:3] "Phylum" "Family" "Genus"
## ..@ sam_data :'data.frame': 1151 obs. of 10 variables:
## Formal class 'sample_data' [package "phyloseq"] with 4 slots
## .. .. ..@ .Data :List of 10
## .. .. .. ..$ : int [1:1151] 28 24 52 22 25 42 25 27 21 25 ...
## .. .. .. ..$ : Factor w/ 2 levels "female","male": 2 1 2 1 1 2 1 1 1 1 ...
## .. .. .. ..$ : Factor w/ 6 levels "CentralEurope",..: 6 6 6 6 6 6 6 6 6 6 ...
## .. .. .. ..$ : Factor w/ 3 levels "o","p","r": NA NA NA NA NA NA NA NA NA NA ...
## .. .. .. ..$ : Factor w/ 40 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
## .. .. .. ..$ : num [1:1151] 5.76 6.06 5.5 5.87 5.89 5.53 5.49 5.38 5.34 5.64 ...
## .. .. .. ..$ : Factor w/ 6 levels "underweight",..: 5 4 2 1 2 2 1 2 2 2 ...
## .. .. .. ..$ : Factor w/ 1006 levels "1","2","3","4",..: 1 2 3 4 5 6 7 8 9 10 ...
## .. .. .. ..$ : num [1:1151] 0 0 0 0 0 0 0 0 0 0 ...
## .. .. .. ..$ : chr [1:1151] "Sample-1" "Sample-2" "Sample-3" "Sample-4" ...
## .. .. ..@ names : chr [1:10] "age" "sex" "nationality" "DNA_extraction_method" ...
## .. .. ..@ row.names: chr [1:1151] "Sample-1" "Sample-2" "Sample-3" "Sample-4" ...
## .. .. ..@ .S3Class : chr "data.frame"
## ..@ phy_tree : NULL
## ..@ refseq : NULLPull out the data frame that contains information on each study subject in the atlas1006 data by using the get_variable accessor function:
## age sex nationality DNA_extraction_method project diversity
## Sample-1 28 male US <NA> 1 5.76
## Sample-2 24 female US <NA> 1 6.06
## Sample-3 52 male US <NA> 1 5.50
## Sample-4 22 female US <NA> 1 5.87
## Sample-5 25 female US <NA> 1 5.89
## Sample-6 42 male US <NA> 1 5.53
## bmi_group subject time sample
## Sample-1 severeobese 1 0 Sample-1
## Sample-2 obese 2 0 Sample-2
## Sample-3 lean 3 0 Sample-3
## Sample-4 underweight 4 0 Sample-4
## Sample-5 lean 5 0 Sample-5
## Sample-6 lean 6 0 Sample-6Note: Since the first argument to get_variable is the phyloseq object (here, the atlas1006 data object), you can pipe into the function, like this:
Find out the different nationalities included in the data:
atlas1006 %>%
get_variable() %>%
as_tibble() %>% # Change output from a data.frame to a tibble
group_by(nationality) %>%
count()## # A tibble: 7 × 2
## # Groups: nationality [7]
## nationality n
## <fct> <int>
## 1 CentralEurope 650
## 2 EasternEurope 15
## 3 Scandinavia 271
## 4 SouthEurope 89
## 5 UKIE 50
## 6 US 44
## 7 <NA> 32It looks like most subjects were from Central Europe, with the next-largest group from
Scandinavia. Note: If you wanted to rearrange this summary to give the nationalities
in order of the number of subjects in each, you could add on a forcats function:
atlas1006 %>%
get_variable() %>%
as_tibble() %>% # Change output from a data.frame to a tibble
mutate(nationality = fct_infreq(nationality)) %>%
group_by(nationality) %>%
count() ## # A tibble: 7 × 2
## # Groups: nationality [7]
## nationality n
## <fct> <int>
## 1 CentralEurope 650
## 2 Scandinavia 271
## 3 SouthEurope 89
## 4 UKIE 50
## 5 US 44
## 6 EasternEurope 15
## 7 <NA> 32Find out the different BMI groups included in the data and if the study seemed to be balanced across those groups:
atlas1006 %>%
get_variable() %>%
as_tibble() %>% # Change output from a data.frame to a tibble
group_by(bmi_group) %>%
count()## # A tibble: 7 × 2
## # Groups: bmi_group [7]
## bmi_group n
## <fct> <int>
## 1 underweight 21
## 2 lean 484
## 3 overweight 197
## 4 obese 222
## 5 severeobese 99
## 6 morbidobese 22
## 7 <NA> 106There were six different BMI groups. Over 100 study subjects had this information missing. The samples were not evenly distributed across these BMI groups. Instead, the most common (lean) had almost 500 subjects, while the smaller BMI-group samples were around 20 people.
See if it looks like diversity varies much between males and females:
atlas1006 %>%
get_variable() %>%
as_tibble() %>%
group_by(sex) %>%
summarize(mean_diversity = mean(diversity))## # A tibble: 3 × 2
## sex mean_diversity
## <fct> <dbl>
## 1 female 5.84
## 2 male 5.85
## 3 <NA> 5.76See if it looks like diversity varies much across BMI groups:
atlas1006 %>%
get_variable() %>%
as_tibble() %>%
group_by(bmi_group) %>%
summarize(mean_diversity = mean(diversity))## # A tibble: 7 × 2
## bmi_group mean_diversity
## <fct> <dbl>
## 1 underweight 5.84
## 2 lean 5.89
## 3 overweight 5.83
## 4 obese 5.82
## 5 severeobese 5.74
## 6 morbidobese 5.67
## 7 <NA> 5.81Here is the code for the plot:
library(tidyverse)
library(ggthemes)
library(microbiome)
data(atlas1006)
atlas1006 %>%
get_variable() %>%
ggplot(aes(x = bmi_group, y = diversity, color = sex)) +
geom_boxplot(alpha = 0.2) +
facet_wrap(~ nationality)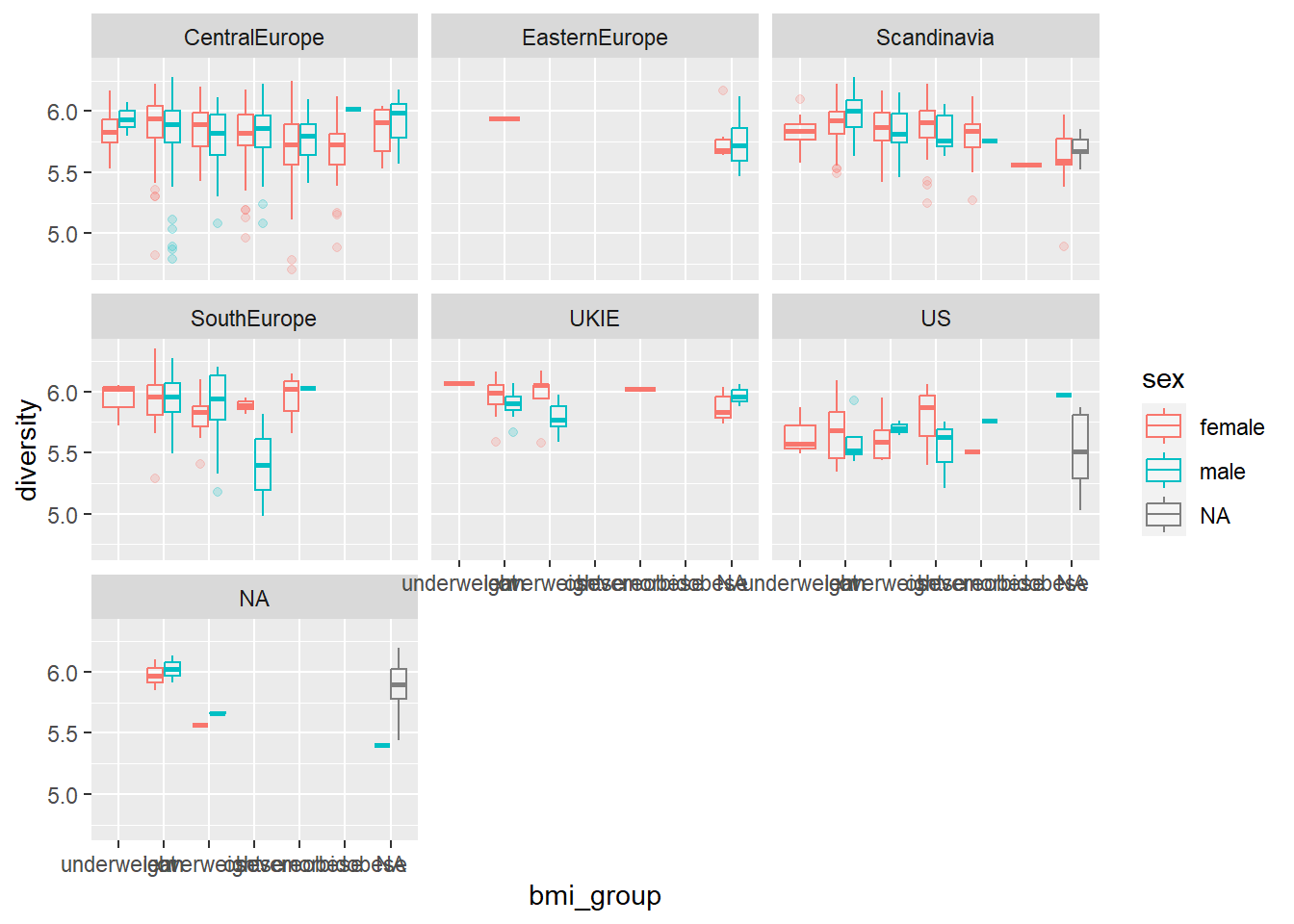
7.8.2 More with baby names
Let’s look at baby names that we started looking at last class, based on the letter they start with.
- For the full dataframe, what proportion of baby names start with each letter? See if you can create a figure to help show this. Create the same plot using the names of people from our class.
- What proportion of names start with “C” or “S” across the full dataset?
7.8.2.1 Example R code
For the full dataframe, what proportion of baby names start with “S”?
To start, create a column with the first letter of each name. You can use
functions in the stringr package to do this. The easiest might be to
use the position of the first letter to pull that information.
library(babynames)
library(stringr)
top_letters <- babynames %>%
mutate(first_letter = str_sub(name, 1, 1))
top_letters %>%
select(name, first_letter) %>%
slice(1:5)## # A tibble: 5 × 2
## name first_letter
## <chr> <chr>
## 1 Mary M
## 2 Anna A
## 3 Emma E
## 4 Elizabeth E
## 5 Minnie MNow we can group by letter and figure out these proportions. First, while the data is grouped, count the number of names with each letter. Then, ungroup and use mutate to divide this by the total number of names:
top_letters <- top_letters %>%
group_by(first_letter) %>%
summarize(n = sum(n)) %>%
ungroup() %>%
mutate(prop = n / sum(n)) %>%
arrange(desc(prop))
top_letters## # A tibble: 26 × 3
## first_letter n prop
## <chr> <int> <dbl>
## 1 J 44612175 0.128
## 2 M 32864210 0.0944
## 3 A 28855232 0.0829
## 4 C 25533863 0.0733
## 5 D 24240271 0.0696
## 6 R 23702794 0.0681
## 7 S 21373830 0.0614
## 8 L 18942067 0.0544
## 9 E 17033760 0.0489
## 10 K 17006684 0.0489
## # ℹ 16 more rowsHere’s one way to visualize this:
library(scales)
top_letters %>%
mutate(first_letter = fct_reorder(first_letter, prop)) %>%
ggplot(aes(x = first_letter)) +
geom_bar(aes(weight = prop)) +
coord_flip() +
scale_y_continuous(labels = percent) +
labs(x = "", y = "Percent of names that start with ...")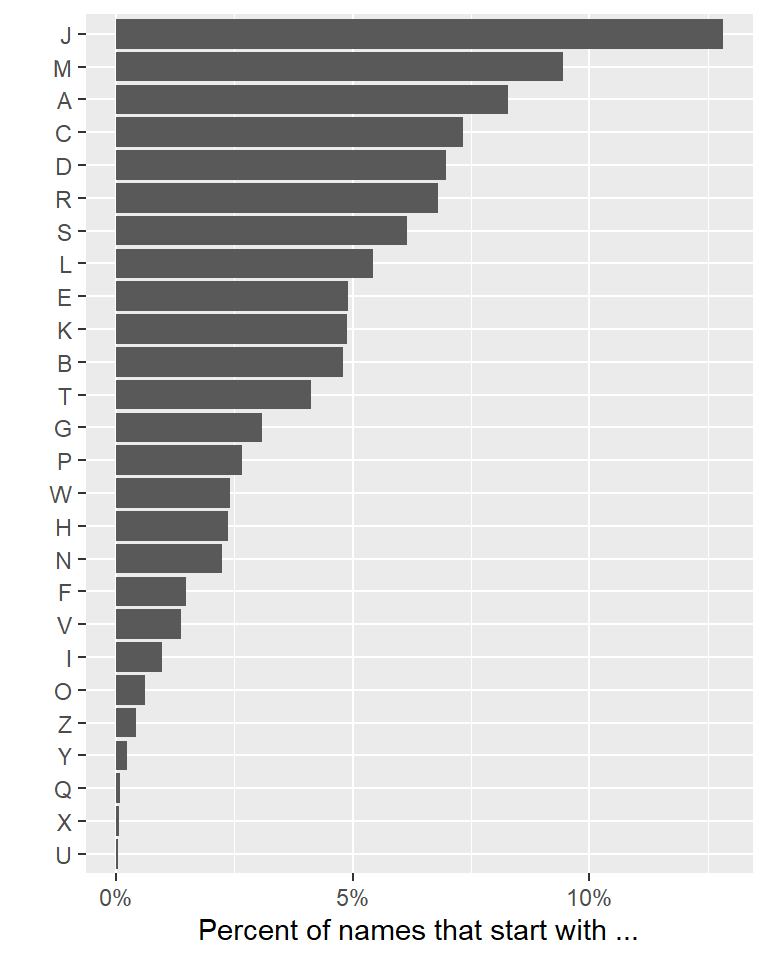
Create the same plot using the names of people in our class. First, create a vector with the names of people in our class:
student_list <- data_frame(name = c("Burton", "Caroline", "Chaoyu", "Collin",
"Daniel", "Eric", "Erin", "Heather",
"Jacob", "Jessica", "Khum", "Kyle",
"Matthew", "Molly", "Nikki", "Rachel",
"Sere", "Shayna", "Sherry", "Sunny"))
student_list <- student_list %>%
mutate(first_letter = str_sub(name, 1, 1))
student_list## # A tibble: 20 × 2
## name first_letter
## <chr> <chr>
## 1 Burton B
## 2 Caroline C
## 3 Chaoyu C
## 4 Collin C
## 5 Daniel D
## 6 Eric E
## 7 Erin E
## 8 Heather H
## 9 Jacob J
## 10 Jessica J
## 11 Khum K
## 12 Kyle K
## 13 Matthew M
## 14 Molly M
## 15 Nikki N
## 16 Rachel R
## 17 Sere S
## 18 Shayna S
## 19 Sherry S
## 20 Sunny Slibrary(scales)
student_list %>%
group_by(first_letter) %>%
count() %>%
ungroup() %>%
mutate(prop = n / sum(n)) %>%
mutate(first_letter = fct_reorder(first_letter, prop)) %>%
ggplot(aes(x = first_letter)) +
geom_bar(aes(weight = prop)) +
coord_flip() +
scale_y_continuous(labels = percent) +
labs(x = "", y = "Percent of students with\na name that starts with ...")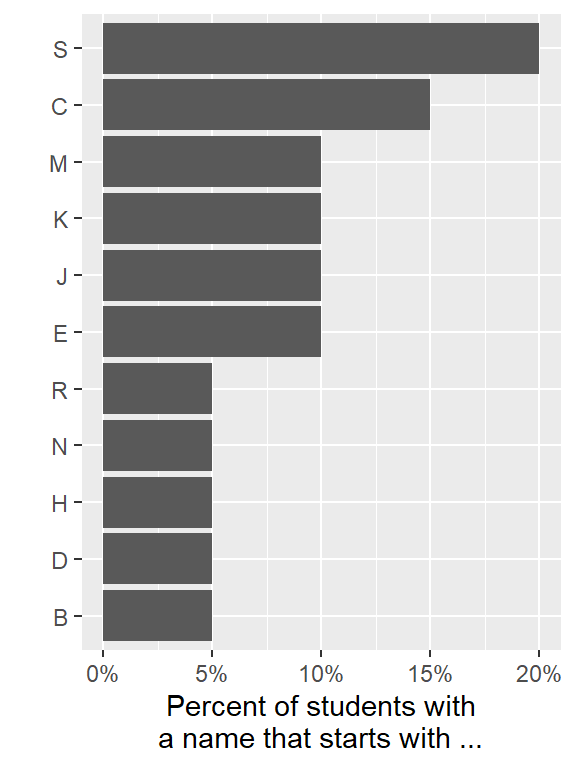
What proportion of names start with “C” or “S” across the full dataset? You can create a dataframe that (1) pulls out the first letter of each name (just like we did for the last part of the question) and (2) tests whether that first letter is an “A” or a “K” (using a logical statement):
c_or_s <- babynames %>%
mutate(first_letter = str_sub(name, 1, 1),
c_or_s = first_letter %in% c("C", "S"))
c_or_s %>%
select(name, first_letter, c_or_s) %>%
slice(1:5)## # A tibble: 5 × 3
## name first_letter c_or_s
## <chr> <chr> <lgl>
## 1 Mary M FALSE
## 2 Anna A FALSE
## 3 Emma E FALSE
## 4 Elizabeth E FALSE
## 5 Minnie M FALSENext, group by this logical column (c_or_s) and figure out the number of
baby names for each group. Then, to get the proportion of the total, ungroup
and mutate to divide by the total number across the data:
## # A tibble: 2 × 3
## c_or_s n prop
## <lgl> <int> <dbl>
## 1 FALSE 1655755 0.860
## 2 TRUE 268910 0.1407.8.3 Running a simple statistical test
In the last part of the in-course exercise, we found out that about 14% of babies born in the United States between 1980 and 1995 had names that started with an “C” or “S” (268,910 babies out of 1,924,665).
- What is the proportion of people with names that start with an “C” or “S” in our class?
- Use a simple statistical test to test the hypothesis that the class comes from a binomial distribution with the same distribution as babies born in the US over the time tracked by this data, in terms of chance of having a name that starts with “C” or “S”. (Hint: You will be comparing two proportions. Try googling for a statistical test in R that does that.)
- See if you can figure out a way to make a single “tidy” pipeline for the whole analysis (and output the result as a tidy dataframe). Does the
tidyfunction frombroomgive different information from this test than the output we got for the Shapiro-Wilk test? - You may get the warning “Chi-squared approximation may be incorrect”. See if you can figure out this warning if you get it with the test you used.
7.8.3.1 Example R code
Here is a vector with names in our class:
library(stringr)
student_list <- tibble(name = c("Burton", "Caroline", "Chaoyu", "Collin",
"Daniel", "Eric", "Erin", "Heather",
"Jacob", "Jessica", "Khum", "Kyle",
"Matthew", "Molly", "Nikki", "Rachel",
"Sere", "Shayna", "Sherry", "Sunny"))
student_list <- student_list %>%
mutate(first_letter = str_sub(name, 1, 1))
student_list %>%
slice(1:3)## # A tibble: 3 × 2
## name first_letter
## <chr> <chr>
## 1 Burton B
## 2 Caroline C
## 3 Chaoyu CLet’s get the total number of students, and then the total number with a name that starts with “C” or “S”:
## # A tibble: 1 × 1
## n
## <int>
## 1 20c_or_s_students <- student_list %>%
mutate(c_or_s = first_letter %in% c("C", "S")) %>%
group_by(c_or_s) %>%
count()
c_or_s_students## # A tibble: 2 × 2
## # Groups: c_or_s [2]
## c_or_s n
## <lgl> <int>
## 1 FALSE 13
## 2 TRUE 7The proportion of students with names starting with “A” or “K” are 7 / 20 = 0.35.
You could run a statistical test comparing these two proportions (check the help file for the function to figure out where to include each piece):
## Warning in prop.test(x = c(268910, 7), n = c(1924665, 20)): Chi-squared
## approximation may be incorrect##
## 2-sample test for equality of proportions with continuity correction
##
## data: c(268910, 7) out of c(1924665, 20)
## X-squared = 5.7121, df = 1, p-value = 0.01685
## alternative hypothesis: two.sided
## 95 percent confidence interval:
## -0.44432032 0.02375596
## sample estimates:
## prop 1 prop 2
## 0.1397178 0.3500000There are a few different ways you could run this test. For example, you could also test whether the proportion in our class is consistent with the null hypothesis that you were drawn from a binomial distribution with a proportion of 0.14 (in-line with the national values):
## Warning in prop.test(x = 7, n = 20, p = 0.14): Chi-squared approximation may be
## incorrect##
## 1-sample proportions test with continuity correction
##
## data: 7 out of 20, null probability 0.14
## X-squared = 5.6852, df = 1, p-value = 0.01711
## alternative hypothesis: true p is not equal to 0.14
## 95 percent confidence interval:
## 0.1630867 0.5905104
## sample estimates:
## p
## 0.35You can also see if we can pipe into prop.test by summing up the number of successes (“1”: the person’s name starts with “C” or “S”). Because this is using an unsummarized
form of the data, it lets us use some of the tidyverse tools more easily:
library(purrr)
library(broom)
student_list %>%
mutate(c_or_s = first_letter %in% c("C", "S")) %>%
pull("c_or_s") %>%
sum() %>%
prop.test(n = 20, p = 0.14) %>%
tidy()## Warning in prop.test(., n = 20, p = 0.14): Chi-squared approximation may be
## incorrect## # A tibble: 1 × 8
## estimate statistic p.value parameter conf.low conf.high method alternative
## <dbl> <dbl> <dbl> <int> <dbl> <dbl> <chr> <chr>
## 1 0.35 5.69 0.0171 1 0.163 0.591 1-sample … two.sidedFinally, when we run this test, we get the warning that “Chi-squared approximation may be incorrect”. Based on googling ‘r prop.test “Chi-squared approximation may be incorrect”’,
it sounds like we might be getting this error because we have a pretty low number of
people in the class. One recommendation is to use binom.test, which will run as an exact binomial test:
##
## Exact binomial test
##
## data: 7 and 20
## number of successes = 7, number of trials = 20, p-value = 0.01534
## alternative hypothesis: true probability of success is not equal to 0.14
## 95 percent confidence interval:
## 0.1539092 0.5921885
## sample estimates:
## probability of success
## 0.357.8.4 Using regression models to explore data #1
For this exercise, you will need the following packages. If do not have them already, you will need to install them.
For this part of the exercise, you’ll use a dataset on weather, air pollution, and mortality counts in Chicago, IL. This dataset is called chicagoNMMAPS and is part of the dlnm package. Change the name of the data frame to chic (this object name is shorter and will be easier to work with). Check out the data a bit to see what variables you have, and then perform the following tasks:
- Write out (on paper, not in R) the regression equation for regressing dewpoint temperature on temperature.
- Try fitting a linear regression of dew point temperature (
dptp) regressed on temperature (temp). Save this model as the objectmod_1(i.e., is the dependent variable of dewpoint temperature linearly associated with the independent variable of temperature). - Based on this regression, does there seem to be a relationship between temperature and dewpoint temperature in Chicago? (Hint: Try using
glanceandtidyfrom thebroompackage on the model object to get more information about the model you fit.) What is the coefficient for temperature (in other words, for every 1 degree increase in temperature, how much do we expect the dewpoint temperature to change?)? What is the p-value for the coefficient for temperature? - Plot temperature (x-axis) versus dewpoint temperature (y-axis) for Chicago. Add in the regression line from the model you fit by using the results from
augment. - Use
autoploton the model object to generate some model diagnostic plots (make sure you have theggfortifypackage loaded and installed).
7.8.4.1 Example R code:
The regression equation for the model you want to fit, regressing dewpoint temperature on temperature, is:
\[ Y_t \sim \beta_0 + \beta_1 X_t \]
where \(Y_t\) is the dewpoint temperature on day \(t\), \(X_t\) is the temperature on day \(t\), and \(\beta_0\) and \(\beta_1\) are model coefficients.
Install and load the dlnm package and then load the chicagoNMMAPS data. Change the name of the data frame to chic, so it will be shorter to call for the rest of your work.
Fit a linear regression of dptp on temp and save as the object mod_1:
##
## Call:
## lm(formula = dptp ~ temp, data = chic)
##
## Coefficients:
## (Intercept) temp
## 24.025 1.621Use functions from the broom package to pull the same information about the model in a “tidy” format. To find out if the evidence for a linear association between temperature and dewpoint temperature, use the tidy function to get model coefficients in a tidy format:
## # A tibble: 2 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 24.0 0.113 213. 0
## 2 temp 1.62 0.00763 212. 0There does seem to be an association between temperature and dewpoint temperature: a unit increase in temperature is associated with a 1.6 unit increase in dewpoint temperature. The p-value for the temperature coefficient is <2e-16. This is far below 0.05, which suggests we would be very unlikely to see such a strong association by chance if the null hypothesis, that the two variables are not associated, were true.
You can also check overall model summaries using the glance function:
## # A tibble: 1 × 12
## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.898 0.898 5.90 45108. 0 1 -16332. 32670. 32690.
## # ℹ 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>To create plots of the observations and the fit model, use the augment function to add model output (e.g., predictions, residuals) to the original data frame of observed temperatures and dew point temperatures:
## # A tibble: 3 × 8
## dptp temp .fitted .resid .hat .sigma .cooksd .std.resid
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 31.5 -0.278 23.6 7.93 0.000376 5.90 0.000340 1.34
## 2 29.9 0.556 24.9 4.95 0.000348 5.90 0.000123 0.839
## 3 27.4 0.556 24.9 2.45 0.000348 5.90 0.0000300 0.415Plot these two variables and add in the fitted line from the model (note: I’ve used the color option to make the color of the points gray). Use the output from augment to create a plot of the original data, with the predicted values used to plot a fitted line.
augment(mod_1) %>%
ggplot(aes(x = temp, y = dptp)) +
geom_point(size = 0.8, alpha = 0.5, col = "gray") +
geom_line(aes(x = temp, y = .fitted), color = "red", size = 2) +
theme_classic()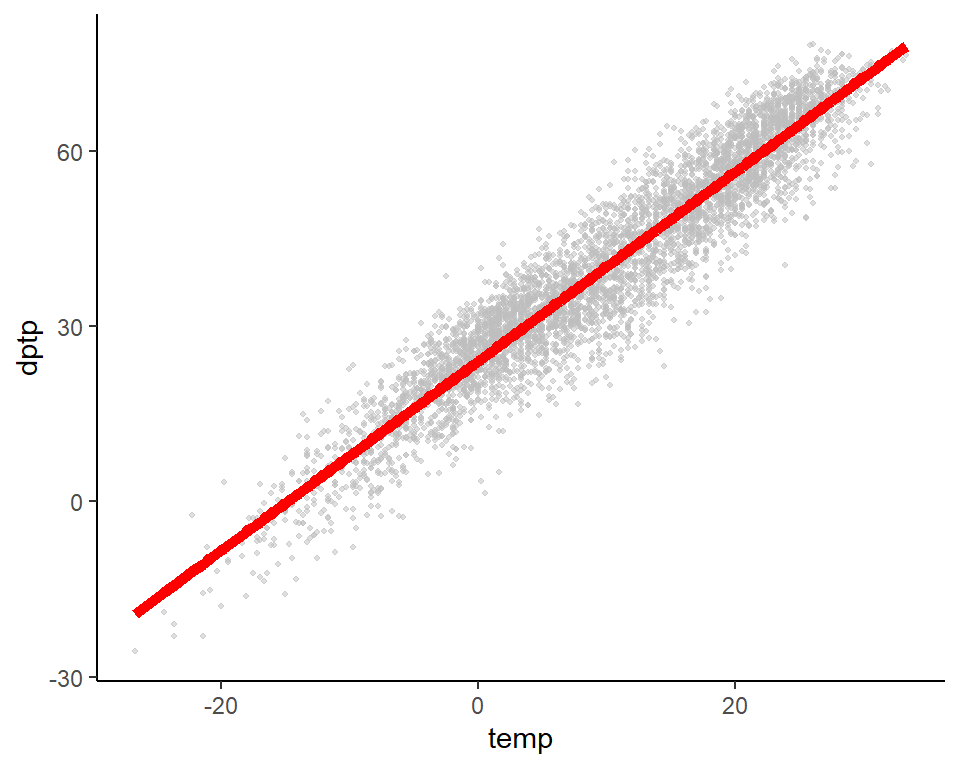
Plot some plots to check model assumptions for the model you fit using the autoplot function on your model object:
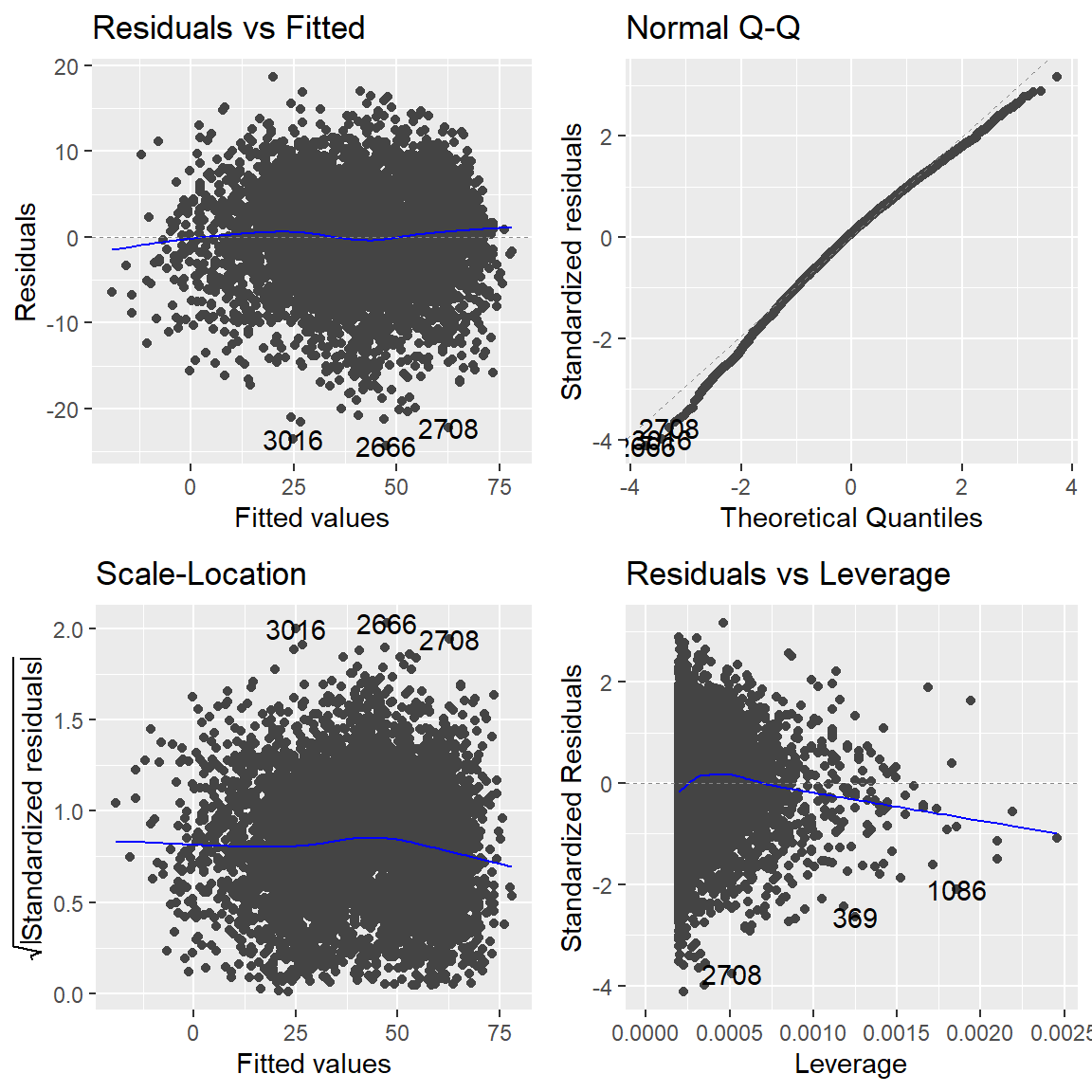
7.8.5 Using regression models to explore data #2
- Try fitting the regression from the last part of the in-course exercise as a GLM, using
glm()(but still assuming the outcome variable is normally distributed). Are your coefficients different? - Does \(PM_{10}\) vary by day of the week? (Hint: The
dowvariable is a factor that gives day of the week. You can do an ANOVA analysis by fitting a linear model using this variable as the independent variable. Some of the overall model summaries will compare this model to an intercept-only model.) What day of the week is \(PM_{10}\) generally highest? (Check the model coefficients to figure this out.) Try to write out (on paper) the regression equation for the model you’re fitting. - Try using
glm()to run a Poisson regression of respiratory deaths (resp) on temperature during summer days. Start by creating a subset with just summer days calledsummer. (Hint: Use themonthfunction with the argumentlabel = TRUEfromlubridateto do this—just pull out the subset where the month June, July, or August.) Try to write out the regression equation for the model you’re fitting. - The coefficient for the temperature variable in this model is our best estimate (based on this model) of the log relative risk for a one degree Celcius increase in temperature. What is the relative risk associated with a one degree Celsius increase?
7.8.5.1 Example R code:
Try fitting the model from the last part of the in-course exercise using glm(). Call it mod_1a. Compare the coefficients for the two models. You can use the tidy function on an lm or glm object to pull out just the model coefficients and associated model results. Here, I’ve used a pipeline of code to create a tidy data frame that merges these “tidy” coefficient outputs (from the two models) into a single data frame):
mod_1a <- glm(dptp ~ temp, data = chic)
tidy(mod_1) %>%
select(term, estimate) %>%
inner_join(mod_1a %>% tidy() %>% select(term, estimate), by = "term") %>%
rename(estimate_lm_mod = estimate.x,
estimate_glm_mod = estimate.y)## # A tibble: 2 × 3
## term estimate_lm_mod estimate_glm_mod
## <chr> <dbl> <dbl>
## 1 (Intercept) 24.0 24.0
## 2 temp 1.62 1.62The results from the two models are identical.
As a note, you could have also just run tidy on each model object, without merging them together into a single data frame:
## # A tibble: 2 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 24.0 0.113 213. 0
## 2 temp 1.62 0.00763 212. 0## # A tibble: 2 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 24.0 0.113 213. 0
## 2 temp 1.62 0.00763 212. 0Fit a model of \(PM_{10}\) regressed on day of week, where day of week is a factor.
## # A tibble: 7 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 27.5 0.730 37.7 7.47e-273
## 2 dowMonday 6.13 1.03 5.93 3.22e- 9
## 3 dowTuesday 6.80 1.03 6.62 4.05e- 11
## 4 dowWednesday 8.48 1.03 8.26 1.85e- 16
## 5 dowThursday 8.80 1.02 8.60 1.08e- 17
## 6 dowFriday 9.48 1.03 9.24 3.61e- 20
## 7 dowSaturday 3.66 1.03 3.56 3.68e- 4Use glance to check some of the overall summaries of this model. The statistic column here is the F statistic from test comparing this model to an intercept-only model.
## # A tibble: 1 × 12
## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.0259 0.0247 19.1 21.5 4.61e-25 6 -21234. 42484. 42536.
## # ℹ 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>As a note, you may have heard in previous statistics classes that you can use the anova() command to compare this model to a model with only an intercept (i.e., one that only fits a global mean and uses that as the expected value for all of the observations). Note that, in this case, the F value from anova for this model comparison is the same as the statistic you got in the overall summary statistics you get with glance in the previous code.
## Analysis of Variance Table
##
## Response: pm10
## Df Sum Sq Mean Sq F value Pr(>F)
## dow 6 46924 7820.6 21.5 < 2.2e-16 ***
## Residuals 4856 1766407 363.8
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The overall p-value from anova for with day-of-week coefficients versus the model that just has an intercept is < 2.2e-16. This is well below 0.05, which suggests that day-of-week is associated with PM10 concentration, as a model that includes day-of-week does a much better job of explaining variation in PM10 than a model without it does.
Use a boxplot to visually compare PM10 by day of week.
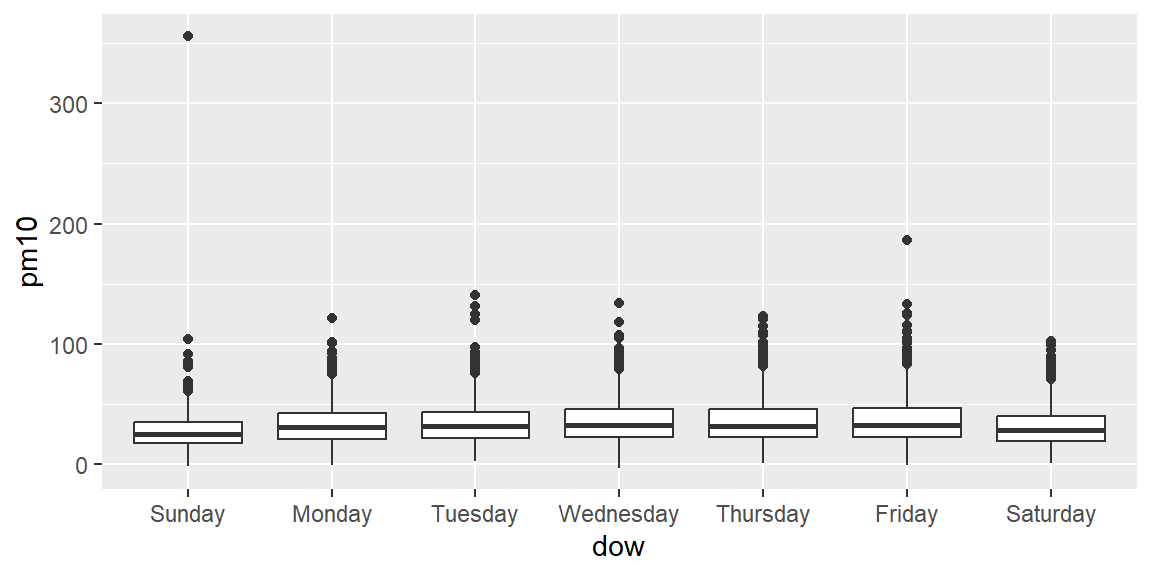
Now try the same plot, but try using the ylim = option to change the limits on the y-axis for the graph, so you can get a better idea of the pattern by day of week (some of the extreme values are very high, which makes it hard to compare by eye when the y-axis extends to include them all).
## Warning: Removed 292 rows containing non-finite outside the scale range
## (`stat_boxplot()`).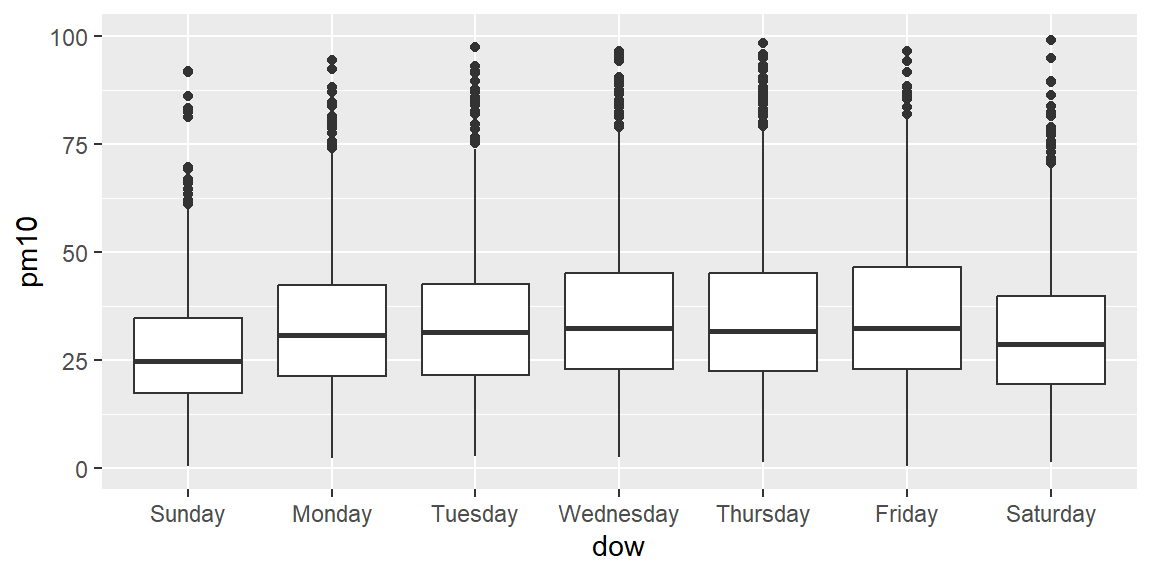
Create a subset called summer with just the summer days:
library(lubridate)
summer <- chic %>%
mutate(month = month(date, label = TRUE)) %>%
filter(month %in% c("Jun", "Jul", "Aug"))
summer %>%
slice(1:3)## date time year month doy dow death cvd resp temp dptp rhum
## 1 1987-06-01 152 1987 Jun 152 Monday 112 60 5 23.61111 68.50 71.875
## 2 1987-06-02 153 1987 Jun 153 Tuesday 111 57 7 22.22222 64.75 95.250
## 3 1987-06-03 154 1987 Jun 154 Wednesday 120 59 9 20.55556 47.25 47.125
## pm10 o3
## 1 22.95607 34.94623
## 2 31.31339 18.96620
## 3 34.95607 23.59270Use glm() to fit a Poisson model of respiratory deaths regressed on temperature. Since you want to fit a Poisson model, use the option family = poisson(link = "log").
## # A tibble: 1 × 8
## null.deviance df.null logLik AIC BIC deviance df.residual nobs
## <dbl> <int> <dbl> <dbl> <dbl> <dbl> <int> <int>
## 1 1499. 1287 -3211. 6425. 6436. 1494. 1286 1288## # A tibble: 2 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 1.91 0.0584 32.7 6.60e-235
## 2 temp 0.00614 0.00258 2.38 1.74e- 2Use the fitted model coefficient to determine the relative risk for a one degree Celsius increase in temperature. First, remember that you can use the tidy() function to read out the model coefficients. The second of these is the value for the temperature coefficient. That means that you can use indexing ([2]) to get just that value. That’s the log relative risk; take the exponent to get the relative risk.
## # A tibble: 1 × 6
## term estimate std.error statistic p.value log_rr
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 temp 0.00614 0.00258 2.38 0.0174 1.01As a note, you can use the conf.int parameter in tidy to also pull confidence intervals:
## # A tibble: 2 × 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 1.91 0.0584 32.7 6.60e-235 1.80 2.02
## 2 temp 0.00614 0.00258 2.38 1.74e- 2 0.00108 0.0112You could use this to get the confidence interval for relative risk (check out the mutate_at function if you haven’t seen it before):
tidy(mod_3, conf.int = TRUE) %>%
select(term, estimate, conf.low, conf.high) %>%
filter(term == "temp") %>%
mutate_at(vars(estimate:conf.high), funs(exp(.)))## Warning: `funs()` was deprecated in dplyr 0.8.0.
## ℹ Please use a list of either functions or lambdas:
##
## # Simple named list: list(mean = mean, median = median)
##
## # Auto named with `tibble::lst()`: tibble::lst(mean, median)
##
## # Using lambdas list(~ mean(., trim = .2), ~ median(., na.rm = TRUE))
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.## # A tibble: 1 × 4
## term estimate conf.low conf.high
## <chr> <dbl> <dbl> <dbl>
## 1 temp 1.01 1.00 1.017.8.6 Trying out nesting and mapping
- We’ll be using data that’s in an R package called “nycflights13”. This data package can be installed from CRAN. Install the package and then load the package and its “flights” dataset.
So that this data will be easier to work with, remove all columns except for those for
the departure delay (
dep_delay), the carrier (carrier), the hour the flight was supposed to leave (hour), and the airport the flight left from (origin). Also, limit the dataset to only flights that left from LaGuardia Airport (“LGA”). - We want to figure out if the probability of a flight leaving 15 minutes late or more
increases over the day for flights leaving LaGuardia. Filter out all the rows where the
departure delay is missing and then create a new column called
late_depthat is true if the flight left 15 minutes or more late and false otherwise. What proportion of all flights leaving from LaGuardia leave 15 minutes late or later? - Next, determine what proportion of all flights are delayed base on the hour that the
flight was scheduled to depart (
hour). Create a plot showing how the probability of leaving 15 minutes or more late changes by hour. - Fit a generalized linear model for the association between the binary variable of whether
the flight was 15 minutes or more late (
late_dep) and the hour the flight was scheduled to leave (hour). Use a binomial model (addfamily = binomial(link = "logit")in theglmcall). The estimate from the model forhourwill be an estimate of the log odds ratio for a one-hour increase in scheduled departure time. Take the exponent of this estimate withexpto get an estimated odds ratio for a one-hour increase in scheduled departure time. Is this estimate larger than 1.0? - Next, we want to see if this association is similar across airlines. First, create a
dataframe called
nested_flightswhere theflightsdata is grouped by airline (carrier) and then nest the data, so that there is a “data” list-column where each item is a dataframe of flight delay data for a specific carrier. - Use the
mapfunction frompurrrinside amutatestatement to apply theglmcode you used earlier for the whole dataset, but in this case for the data for each airline character. Then use themapfunction inside amutatestatement again to “tidy” the data. - Clean the data up a bit. Remove the columns for
dataandglm_resultand thenunnestthe dataframe list-column with the tidy version of the model results. Filter to get only the estimates for the “hour” term. Then calculate an odds ratio (or) by taking the exponent (check out theexpfunction) of the original estimate. - The package has a dataframe with the full name of each carrier (
airlines). Join this data into the data you’ve been working with, so you have the full names of airlines. - Finally, create the following plot with each airline’s odds ratio for the change in the chance of a delay per one-hour increase in the scheduled hour of departure:
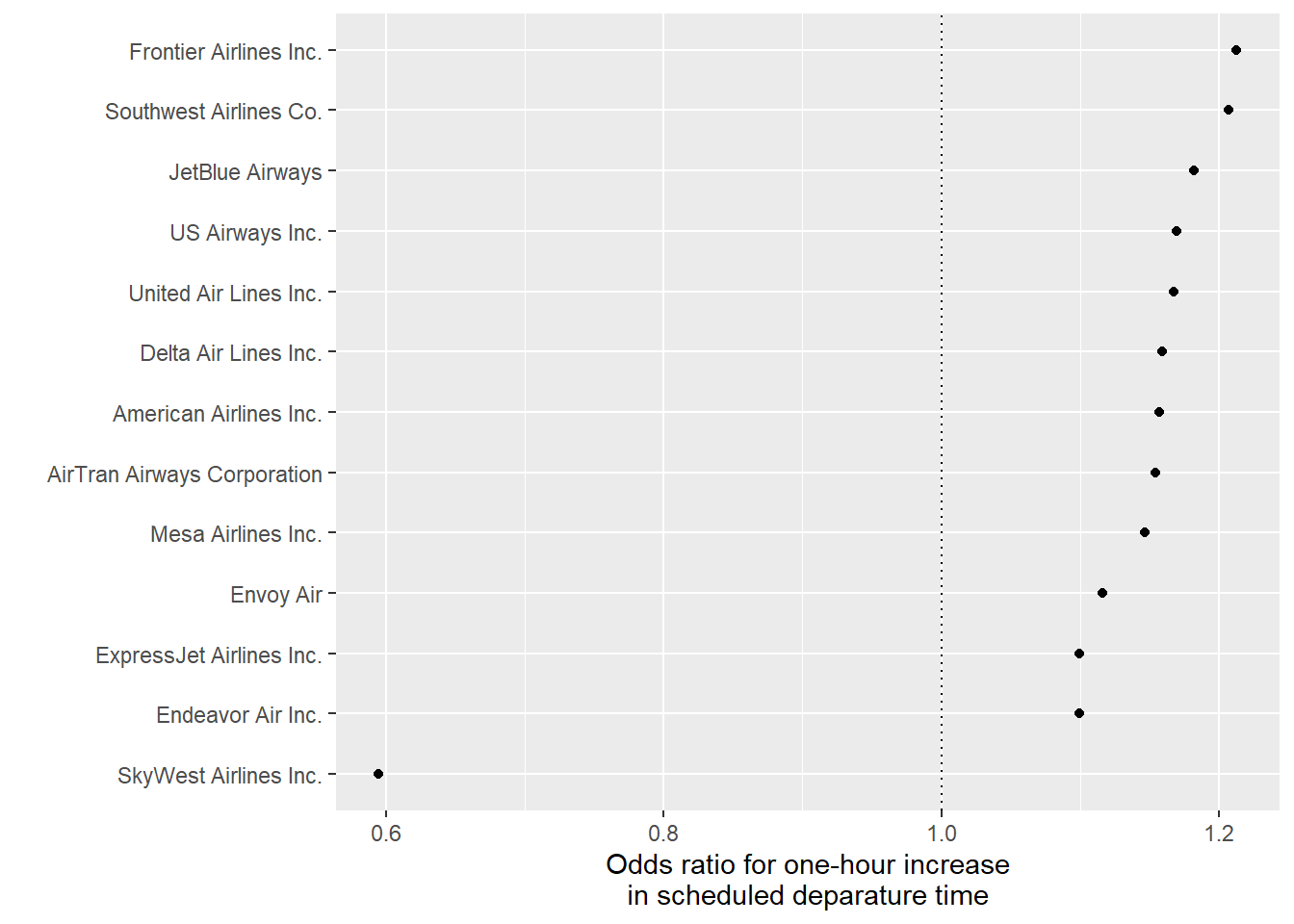
7.8.6.1 Example R code:
Install the “nycflights13” package from CRAN. Load the package and its “flights” dataset.
So that this data will be easier to work with, remove all columns except for those for
the departure dealy (dep_delay), the carrier (carrier), the hour the flight was
supposed to leave (hour), and the airport the flight left from (origin).
## # A tibble: 104,662 × 4
## dep_delay carrier hour origin
## <dbl> <chr> <dbl> <chr>
## 1 4 UA 5 LGA
## 2 -6 DL 6 LGA
## 3 -3 EV 6 LGA
## 4 -2 AA 6 LGA
## 5 -1 AA 6 LGA
## 6 0 B6 6 LGA
## 7 0 MQ 6 LGA
## 8 -8 DL 6 LGA
## 9 -3 MQ 6 LGA
## 10 13 AA 6 LGA
## # ℹ 104,652 more rowsFilter out all the rows where the
departure delay is missing and then create a new column called
late_dep that is true if the flight left 15 minutes or more late and false otherwise.
## # A tibble: 101,509 × 5
## dep_delay carrier hour origin late_dep
## <dbl> <chr> <dbl> <chr> <lgl>
## 1 4 UA 5 LGA FALSE
## 2 -6 DL 6 LGA FALSE
## 3 -3 EV 6 LGA FALSE
## 4 -2 AA 6 LGA FALSE
## 5 -1 AA 6 LGA FALSE
## 6 0 B6 6 LGA FALSE
## 7 0 MQ 6 LGA FALSE
## 8 -8 DL 6 LGA FALSE
## 9 -3 MQ 6 LGA FALSE
## 10 13 AA 6 LGA FALSE
## # ℹ 101,499 more rowsWhat proportion of all flights leaving from LaGuardia leave 15 minutes late or later? To
check this, remember that TRUE is saved as a “1” and FALSE is saved as a “0”. That means that
we can take the mean of a logical vector to get the proportion of trials that are true.
## [1] 0.1889685Determine what proportion of all flights are delayed base on the hour that the
flight was scheduled to depart (hour). Create a plot showing how the probability of
leaving 15 minutes or more late changes by hour.
## # A tibble: 18 × 2
## hour prob_late
## <dbl> <dbl>
## 1 5 0.0588
## 2 6 0.0519
## 3 7 0.0570
## 4 8 0.0967
## 5 9 0.105
## 6 10 0.128
## 7 11 0.159
## 8 12 0.163
## 9 13 0.200
## 10 14 0.207
## 11 15 0.236
## 12 16 0.273
## 13 17 0.292
## 14 18 0.298
## 15 19 0.314
## 16 20 0.331
## 17 21 0.309
## 18 22 0.391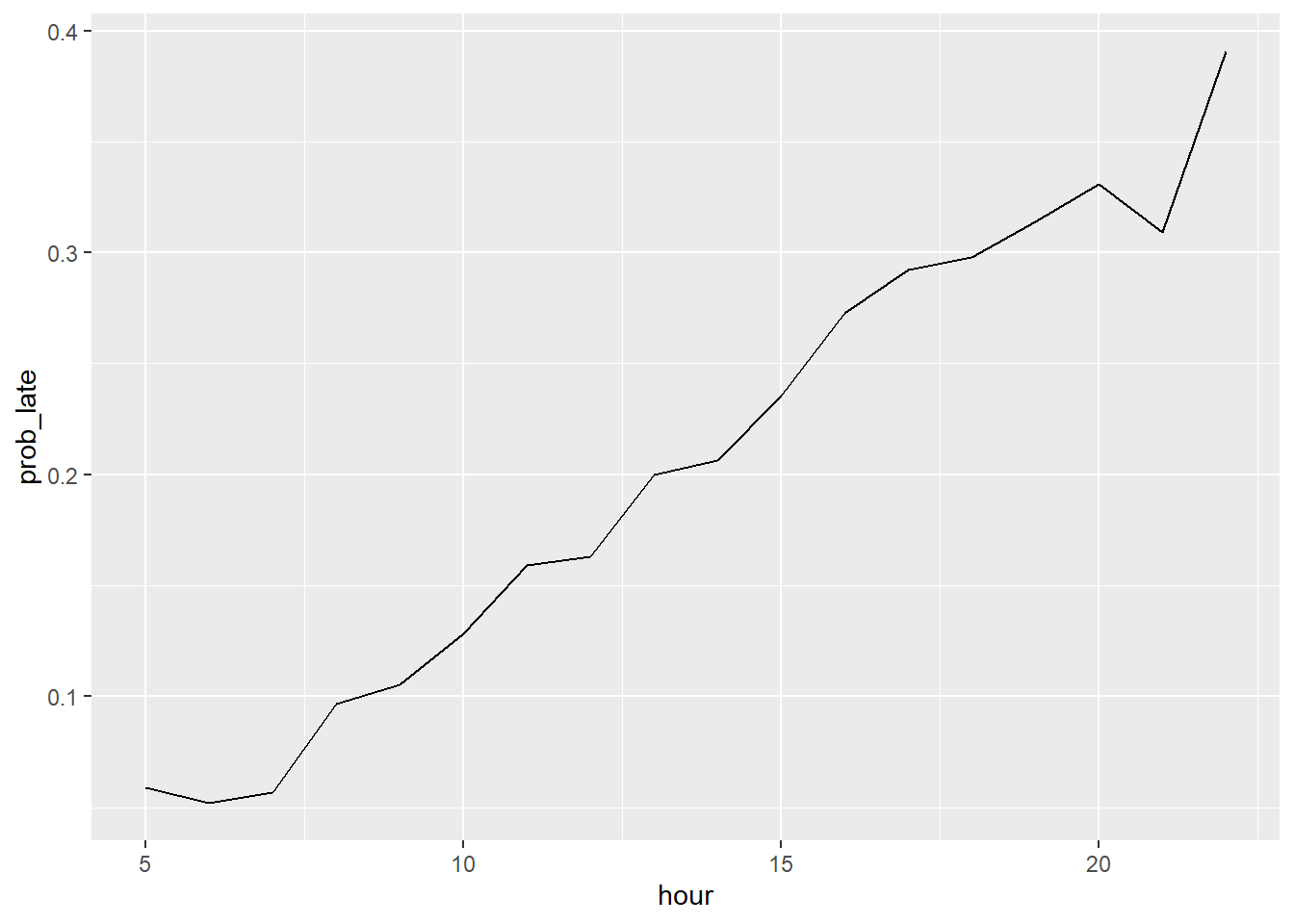
Fit a generalized linear model for the association between the binary variable of whether
the flight was 15 minutes or more late (late_dep) and the hour the flight was
scheduled to leave (hour). Use a binomial model (add family = binomial(link = "logit")
in the glm call). The estimate from the model for hour will be an estimate
of the log odds ratio for a one-hour increase in scheduled departure time. Take the
exponent of this estimate with exp to get an estimated odds ratio for a one-hour
increase in scheduled departure time. Is this estimate larger than 1.0?
##
## Call: glm(formula = late_dep ~ hour, family = binomial(link = "logit"),
## data = flights)
##
## Coefficients:
## (Intercept) hour
## -3.3643 0.1399
##
## Degrees of Freedom: 101508 Total (i.e. Null); 101507 Residual
## Null Deviance: 98410
## Residual Deviance: 92810 AIC: 92810library(broom)
glm(late_dep ~ hour, data = flights, family = binomial(link = "logit")) %>%
tidy() %>% # Tidy the model results
filter(term == "hour") %>% # Only look at the estimate for `hour`
mutate(or = exp(estimate)) # Estimate is log odds ratio. Take exponent for odds ratio## # A tibble: 1 × 6
## term estimate std.error statistic p.value or
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 hour 0.140 0.00195 71.7 0 1.15Next, we want to see if this association is similar across airlines. First, create a
dataframe called nested_flights where the flights
data is grouped by airline (carrier) and then nest the data, so that there is a “data” list-column
where each item is a dataframe of flight delay data for a specific carrier:
## # A tibble: 13 × 2
## # Groups: carrier [13]
## carrier data
## <chr> <list>
## 1 UA <tibble [7,837 × 4]>
## 2 DL <tibble [22,857 × 4]>
## 3 EV <tibble [8,255 × 4]>
## 4 AA <tibble [15,063 × 4]>
## 5 B6 <tibble [5,925 × 4]>
## 6 MQ <tibble [16,189 × 4]>
## 7 WN <tibble [6,000 × 4]>
## 8 FL <tibble [3,187 × 4]>
## 9 US <tibble [12,574 × 4]>
## 10 F9 <tibble [682 × 4]>
## 11 9E <tibble [2,372 × 4]>
## 12 YV <tibble [545 × 4]>
## 13 OO <tibble [23 × 4]>To check the contents of the list-column, try:
## # A tibble: 6 × 4
## dep_delay hour origin late_dep
## <dbl> <dbl> <chr> <lgl>
## 1 4 5 LGA FALSE
## 2 -4 6 LGA FALSE
## 3 1 6 LGA FALSE
## 4 9 7 LGA FALSE
## 5 -4 7 LGA FALSE
## 6 2 7 LGA FALSEUse the map function from purrr inside a mutate statement to apply the glm code you used earlier for the
whole dataset, but in this case for the data for each airline character:
library(purrr)
library(tidyr)
prob_late <- nested_flights %>%
mutate(glm_result = purrr::map(data, ~ glm(late_dep ~ hour,
data = .x, family = binomial(link = "logit"))))
# Check the results for the first element of the "glm_result" column:
prob_late$glm_result[[1]]##
## Call: glm(formula = late_dep ~ hour, family = binomial(link = "logit"),
## data = .x)
##
## Coefficients:
## (Intercept) hour
## -3.4305 0.1551
##
## Degrees of Freedom: 7836 Total (i.e. Null); 7835 Residual
## Null Deviance: 7601
## Residual Deviance: 7064 AIC: 7068Then use the map
function inside a mutate statement again to “tidy” the data.
Remove the columns for data and
glm_result:
Then `unnest the dataframe list-column with the tidy version of the model
results:
Filter to get only the estimates for the “hour” term:
Then calculate an odds ratio
(or) by taking the exponent (check the exp function) of the original estimate.
## # A tibble: 6 × 7
## # Groups: carrier [6]
## carrier term estimate std.error statistic p.value or
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 UA hour 0.155 0.00705 22.0 2.88e-107 1.17
## 2 DL hour 0.148 0.00454 32.6 1.80e-232 1.16
## 3 EV hour 0.0952 0.00572 16.7 2.79e- 62 1.10
## 4 AA hour 0.146 0.00580 25.2 1.05e-139 1.16
## 5 B6 hour 0.168 0.00702 23.9 7.13e-126 1.18
## 6 MQ hour 0.110 0.00468 23.5 2.23e-122 1.12The package has a dataframe with the full name of each carrier (airlines). Join this data
into the data you’ve been working with, so you have the full names of airlines:
Finally, create the following plot with each airline’s association between hour in the day and the chance of a delay:
library(forcats)
data(airlines)
prob_late %>%
ungroup() %>%
mutate(estimate = exp(estimate)) %>%
mutate(name = fct_reorder(name, estimate)) %>%
ggplot(aes(x = estimate, y = name)) +
geom_point() +
geom_vline(xintercept = 1, linetype = 3) +
labs(x = "Odds ratio for one-hour increase in scheduled deparature time", y = "")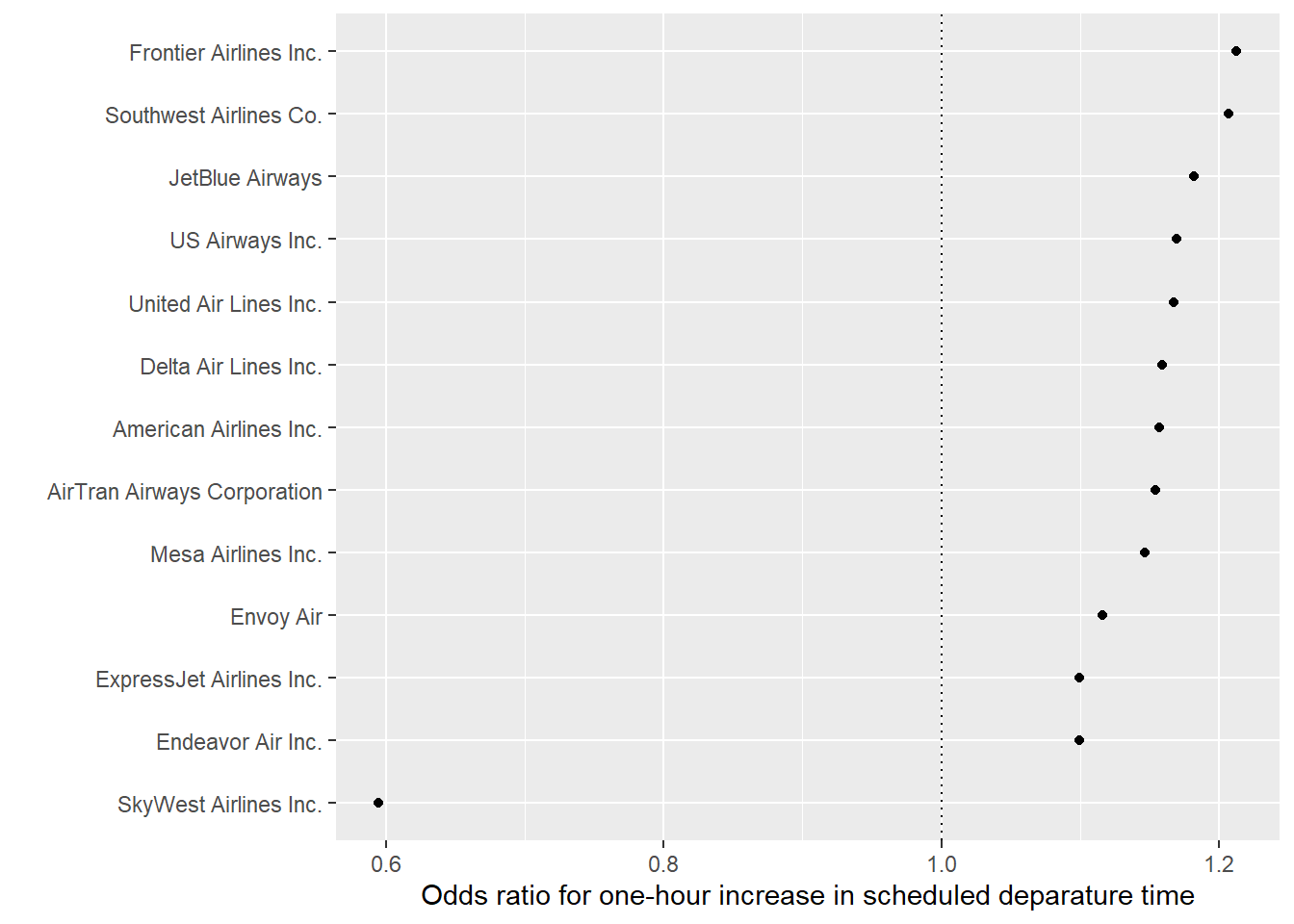
7.8.7 Writing functions
- Say that you have a four-letter character string (e.g., “ling”) and that you want to move the last letter to the front of the string to create a new four-letter character string (e.g., “glin”). Write some R code to do this.
- Next, write a function named
move_letterthat does the same thing—takes a four-letter character string (e.g., “ling”) and creates a new four-letter character string where the last letter in the original string has been moved to the front of the string (e.g., “glin”). It should input one parameter (word, the original four-letter character string or a vector of four-letter character strings). - Read the word list at https://raw.githubusercontent.com/dwyl/english-words/master/words.txt
into an R dataframe called
word_list. It will only have one column; name this columnword. - Write code that can take a vector of character strings (e.g.,
c('ling', 'scat', 'soil')) and return a logical vector that says whether that character string is a word in theword_listdataframe you created in the last step (e.g.,c('FALSE', 'TRUE', 'TRUE')for a vector where the first value isn’t a word but the second and third are). You may find thepullfunction useful in writing this code (to pull thewordcolumn out of theword_listdataframe). - Write a function called
is_wordthat inputs (1) a vector of character strings and (2) a dataframe with a column called “word” that lists real words. The function should return a logical vector saying whether each character string is a real word. The function should have two arguments:words_to_check, which is a vector of character strings (e.g.,c('ling', 'scat', 'soil')), andreal_word_list, which is a dataframe with a column calledwordsof real English words (e.g., theword_listdataframe you created from the word list on GitHub). Set thereal_word_listargument to have the default value ofwords, the dataframe you created earlier in this exercise by reading in the dataframe of English words from GitHub. - Try using the function with a different word list. As an example, you could read in and use the word list of Google’s top 10,000 English words from https://raw.githubusercontent.com/first20hours/google-10000-english/master/google-10000-english-no-swears.txt.
- Try using these functions to solve the word puzzle problem in the last homework.
7.8.7.1 Example R code
Say that you have a four-letter character string (e.g., “ling”) and that you want
to move the last letter to the front of the string to create a new four-letter
character string (e.g., “glin”). You can use functions from the
stringr package to help with this.
library(stringr)
# Start with an example word. You can use the example word from
# the problem statement ('ling').
word <- "ling"
# Break the word into two parts
first_three_letters <- str_sub(word, 1, 3)
last_letter <- str_sub(word, 4, 4)
first_three_letters## [1] "lin"## [1] "g"# Put the parts back together in the right order
new_word <- str_c(last_letter, first_three_letters) # You could also use `paste` or `paste0` here
new_word## [1] "glin"Write a function named move_letter that takes a four-letter character string
(e.g., “ling”) and creates a new four-letter character string where the last
letter in the original string has been moved to the front of the string (e.g.,
“glin”). It should input one parameter (word, the original four-letter character
string or a vector of four-letter character strings).
To do this, take the code that you just wrote and put it inside a function named
move_letter:
move_letter <- function(word){
# Break the word into two parts
first_three_letters <- str_sub(word, 1, 3)
last_letter <- str_sub(word, 4, 4)
# Put the parts back together in the right order
str_c(last_letter, first_three_letters)
}
# Check the function
move_letter(word = "ling")## [1] "glin"## [1] "scat"## [1] "soil"## [1] "scat" "soil"Notice that now word is being used as an argument in the function. It’s as if you
assigned the string or vector of strings that you want to convert to the object
name word, and then you run all the code. The output of the function is the
last expression in the function code (str_c(last_letter, first_three_letters)).
Also, note that you can add comments inside the function with #, just like you can
with other R code.
Read the word list at https://raw.githubusercontent.com/dwyl/english-words/master/words.txt
into an R dataframe called words. It will only have one column; name this column
word_list.
library(readr)
word_list <- read_csv("https://raw.githubusercontent.com/dwyl/english-words/master/words.txt",
col_names = "word")## Warning: One or more parsing issues, call `problems()` on your data frame for details,
## e.g.:
## dat <- vroom(...)
## problems(dat)## Rows: 466550 Columns: 1
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (1): word
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Write code that can take a vector of character strings (e.g., c('ling', 'scat', 'soil'))
and return a logical vector that says whether that character string is a word in the
word_list dataframe you created in the last step (e.g., c('TRUE', 'FALSE', 'TRUE') for
a vector where the second value isn’t a word but the first and third are).
library(purrr)
words_to_check <- c("ling", "scat", "soil")
words_to_check %in% pull(word_list, "word")## [1] TRUE FALSE TRUEThis code works because you can use pull to extract a column (as a vector) from
a dataframe, and then you can use the %in% operator to see if each value in one vector
(the words_to_check vector in this example) is one of the values in the second
vector (the word column from the words dataframe in this example).
Write a function called is_word that inputs (1) a vector of character strings
and (2) a dataframe with a column called “word” that lists real words. The
function should return a logical vector saying whether each character string is
a real word. The function should have two arguments: words_to_check, which is
a vector of character strings and real_word_list which is a dataframe with a
column called words of real English words. Set the real_word_list argument
to have the default value of words, the dataframe you created earlier in this
exercise by reading in the dataframe of English words from GitHub.
# Put the code you wrote inside a function
is_word <- function(words_to_check, real_word_list = word_list){
words_to_check %in% pull(real_word_list, "word")
}
# Check the function
is_word(words_to_check = c("ling", "scat", "soil"))## [1] TRUE FALSE TRUENote that, if you want to use the word_list dataframe for your real word list, you
don’t have to specify that when you call the is_word function, since you
set that as your default value.
If you wanted to use a different word list, you can specify a different value for
the real_word_list argument when you run the function. For example, to use Google’s
top 10,000 English word list from GitHub instead, use:
google_words <- read_csv("https://raw.githubusercontent.com/first20hours/google-10000-english/master/google-10000-english-no-swears.txt",
col_names = "word")## Rows: 9894 Columns: 1
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (1): word
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.## [1] FALSE FALSE TRUEIn this case, there was a rarer word (“ling”) that counted as a word when the original word list was used, but not when the Google top-10,000 words list was used.
Try using these functions to solve the word puzzle problem in the last homework.
word_list %>%
filter(str_detect(word, "^m.{7}$")) %>% # This is using regular expressions
separate(word, into = c("first_word", "second_word"), # Note that you can use `separate` with a number to use position
sep = 4, remove = FALSE) %>%
mutate(second_word = move_letter(second_word)) %>% # Use the first function here
filter(is_word(first_word) & is_word(second_word)) %>% # Use the second function here
unite("new_word", c("first_word", "second_word"), sep = " ") # You can use `unite` to put the two new words in a phrase in one column.## # A tibble: 103 × 2
## word new_word
## <chr> <chr>
## 1 mailings mail sing
## 2 maillots mail slot
## 3 majorate majo erat
## 4 majoring majo grin
## 5 malmiest malm ties
## 6 maltiest malt ties
## 7 mandalas mand sala
## 8 mandated mand date
## 9 mandates mand sate
## 10 mangiest mang ties
## # ℹ 93 more rowsYou can get the choices down to even fewer options if you match the new words against
the Google top-10,000 words list, by using the real_word_list option in the is_word
function you wrote. (However, “maillots” is not a common enough word that you can
also start from that shorter word list!)
word_list %>%
filter(str_detect(word, "^m.{7}$")) %>%
separate(word, into = c("first_word", "second_word"),
sep = 4, remove = FALSE) %>%
mutate(second_word = move_letter(second_word)) %>%
filter(is_word(first_word, real_word_list = google_words) &
is_word(second_word, real_word_list = google_words)) %>%
unite("new_word", c("first_word", "second_word"), sep = " ") ## # A tibble: 43 × 2
## word new_word
## <chr> <chr>
## 1 mailings mail sing
## 2 maillots mail slot
## 3 maintops main stop
## 4 markings mark sing
## 5 maskings mask sing
## 6 massager mass rage
## 7 massages mass sage
## 8 massiest mass ties
## 9 mattings matt sing
## 10 mealiest meal ties
## # ℹ 33 more rows